Blocking the recruitment of naive CD4+ T cells reverses immunosuppression in breast cancer
- PMID: 28290464
- PMCID: PMC5385617
- DOI: 10.1038/cr.2017.34
Blocking the recruitment of naive CD4+ T cells reverses immunosuppression in breast cancer
Abstract
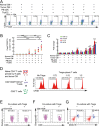
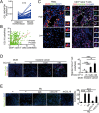
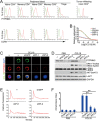
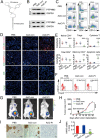
The origin of tumor-infiltrating Tregs, critical mediators of tumor immunosuppression, is unclear. Here, we show that tumor-infiltrating naive CD4+ T cells and Tregs in human breast cancer have overlapping TCR repertoires, while hardly overlap with circulating Tregs, suggesting that intratumoral Tregs mainly develop from naive T cells in situ rather than from recruited Tregs. Furthermore, the abundance of naive CD4+ T cells and Tregs is closely correlated, both indicating poor prognosis for breast cancer patients. Naive CD4+ T cells adhere to tumor slices in proportion to the abundance of CCL18-producing macrophages. Moreover, adoptively transferred human naive CD4+ T cells infiltrate human breast cancer orthotopic xenografts in a CCL18-dependent manner. In human breast cancer xenografts in humanized mice, blocking the recruitment of naive CD4+ T cells into tumor by knocking down the expression of PITPNM3, a CCL18 receptor, significantly reduces intratumoral Tregs and inhibits tumor progression. These findings suggest that breast tumor-infiltrating Tregs arise from chemotaxis of circulating naive CD4+ T cells that differentiate into Tregs in situ. Inhibiting naive CD4+ T cell recruitment into tumors by interfering with PITPNM3 recognition of CCL18 may be an attractive strategy for anticancer immunotherapy.
Figures


References
-
- Nishikawa H, Sakaguchi S. Regulatory T cells in cancer immunotherapy. Curr Opin Immunol 2014; 27:1–7. - PubMed
-
- Beyer M, Schultze JL. Regulatory T cells in cancer. Blood 2006; 108:804–811. - PubMed
-
- Curiel TJ, Coukos G, Zou L, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med 2004; 10:942–949. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

