Nutritional control of IL-23/Th17-mediated autoimmune disease through HO-1/STAT3 activation
- PMID: 28290522
- PMCID: PMC5349589
- DOI: 10.1038/srep44482
Nutritional control of IL-23/Th17-mediated autoimmune disease through HO-1/STAT3 activation
Abstract
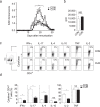
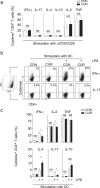
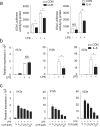
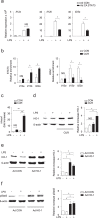
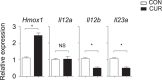
The nutritional curcumin (CUR) is beneficial in cell-mediated autoimmune diseases. The molecular mechanisms underlying this food-mediated silencing of inflammatory immune responses are poorly understood. By investigating antigen-specific immune responses we found that dietary CUR impairs the differentiation of Th1/Th17 cells in vivo during encephalomyelitis and instead promoted Th2 cells. In contrast, feeding CUR had no inhibitory effect on ovalbumin-induced airway inflammation. Mechanistically, we found that CUR induces an anti-inflammatory phenotype in dendritic cells (DC) with enhanced STAT3 phosphorylation and suppressed expression of Il12b and Il23a. On the molecular level CUR readily induced NRF2-sensitive heme oxygenase 1 (HO-1) mRNA and protein in LPS-activated DC. HO-1 enhanced STAT3 phosphorylation, which enriched to Il12b and Il23a loci and negatively regulated their transcription. These findings demonstrate the underlying mechanism through which a nutritional can interfere with the immune response. CUR silences IL-23/Th17-mediated pathology by enhancing HO-1/STAT3 interaction in DC.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
-
- De Rosa V. et al.. Nutritional control of immunity: Balancing the metabolic requirements with an appropriate immune function. Semin Immunol 27, 300–309 (2015). - PubMed
-
- Furusawa Y. et al.. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013). - PubMed
-
- Trompette A. et al.. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 20, 159–166 (2014). - PubMed
-
- Geisel J. et al.. Sulforaphane protects from T cell-mediated autoimmune disease by inhibition of IL-23 and IL-12 in dendritic cells. J Immunol 192, 3530–3539 (2014). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

