Mapping cell surface adhesion by rotation tracking and adhesion footprinting
- PMID: 28290531
- PMCID: PMC5349612
- DOI: 10.1038/srep44502
Mapping cell surface adhesion by rotation tracking and adhesion footprinting
Abstract
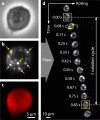
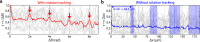
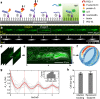
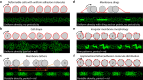
Rolling adhesion, in which cells passively roll along surfaces under shear flow, is a critical process involved in inflammatory responses and cancer metastasis. Surface adhesion properties regulated by adhesion receptors and membrane tethers are critical in understanding cell rolling behavior. Locally, adhesion molecules are distributed at the tips of membrane tethers. However, how functional adhesion properties are globally distributed on the individual cell's surface is unknown. Here, we developed a label-free technique to determine the spatial distribution of adhesive properties on rolling cell surfaces. Using dark-field imaging and particle tracking, we extract the rotational motion of individual rolling cells. The rotational information allows us to construct an adhesion map along the contact circumference of a single cell. To complement this approach, we also developed a fluorescent adhesion footprint assay to record the molecular adhesion events from cell rolling. Applying the combination of the two methods on human promyelocytic leukemia cells, our results surprisingly reveal that adhesion is non-uniformly distributed in patches on the cell surfaces. Our label-free adhesion mapping methods are applicable to the variety of cell types that undergo rolling adhesion and provide a quantitative picture of cell surface adhesion at the functional and molecular level.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

