Abseq: Ultrahigh-throughput single cell protein profiling with droplet microfluidic barcoding
- PMID: 28290550
- PMCID: PMC5349531
- DOI: 10.1038/srep44447
Abseq: Ultrahigh-throughput single cell protein profiling with droplet microfluidic barcoding
Abstract
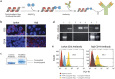
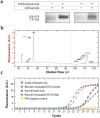
Proteins are the primary effectors of cellular function, including cellular metabolism, structural dynamics, and information processing. However, quantitative characterization of proteins at the single-cell level is challenging due to the tiny amount of protein available. Here, we present Abseq, a method to detect and quantitate proteins in single cells at ultrahigh throughput. Like flow and mass cytometry, Abseq uses specific antibodies to detect epitopes of interest; however, unlike these methods, antibodies are labeled with sequence tags that can be read out with microfluidic barcoding and DNA sequencing. We demonstrate this novel approach by characterizing surface proteins of different cell types at the single-cell level and distinguishing between the cells by their protein expression profiles. DNA-tagged antibodies provide multiple advantages for profiling proteins in single cells, including the ability to amplify low-abundance tags to make them detectable with sequencing, to use molecular indices for quantitative results, and essentially limitless multiplexing.
Conflict of interest statement
The material in this publication is the subject of a patent filing and licensing agreement to Mission Bio, Inc., who is commercializing the technology and of whom ARA is a shareholder.
Figures




References
-
- Gutteridge A. & Thornton J. M. Understanding nature’s catalytic toolkit. Trends Biochem. Sci. 30, 622–629 (2005). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

