Crystal structure of the type IV secretion system component CagX from Helicobacter pylori
- PMID: 28291753
- PMCID: PMC5349311
- DOI: 10.1107/S2053230X17001376
Crystal structure of the type IV secretion system component CagX from Helicobacter pylori
Abstract
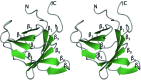
Helicobacter pylori, a Gram-negative bacterial pathogen prevalent in the human population, is the causative agent of severe gastric diseases. An H. pylori type IV secretion (T4S) system encoded by the cytotoxin-associated gene pathogenicity island (cagPAI) is responsible for communication with host cells. As a component of the cagPAI T4S system core complex, CagX plays an important role in virulence-protein translocation into the host cells. In this work, the crystal structure of the C-terminal domain of CagX (CagXct), which is a homologue of the VirB9 protein from the VirB/D4 T4S system, is presented. CagXct is only the second three-dimensional structure to be elucidated of a VirB9-like protein. Another homologue, TraO, which is encoded on the Escherichia coli conjugative plasmid pKM101, shares only 19% sequence identity with CagXct; however, there is a remarkable similarity in tertiary structure between these two β-sandwich protein domains. Most of the residues that are conserved between CagXct and TraO are located within the protein core and appear to be responsible for the preservation of this domain fold. The studies presented here will contribute to our understanding of different bacterial T4S systems.
Keywords: CagX; Helicobacter pylori; X-ray diffraction; cagPAI; crystal structure; cytotoxin-associated gene pathogenicity island; type IV secretion system.
Figures
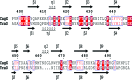
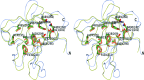
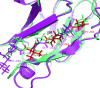
References
-
- Akopyants, N. S., Clifton, S., Kersulyte, D., Crabtree, J. E., Youree, B. E., Reece, C. A., Bukanov, N. O., Drazek, E. S., Roe, B. A. & Berg, D. E. (1998). Mol. Microbiol. 28, 37–53. - PubMed
-
- Backert, S., Churin, Y. & Meyer, T. F. (2002). Keio J. Med. 51, Suppl. 2, 6–14. - PubMed
-
- Backert, S. & Selbach, M. (2008). Cell. Microbiol. 10, 1573–1581. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

