CheckMyMetal: a macromolecular metal-binding validation tool
- PMID: 28291757
- PMCID: PMC5349434
- DOI: 10.1107/S2059798317001061
CheckMyMetal: a macromolecular metal-binding validation tool
Abstract
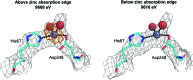
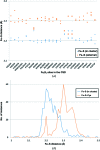
Metals are essential in many biological processes, and metal ions are modeled in roughly 40% of the macromolecular structures in the Protein Data Bank (PDB). However, a significant fraction of these structures contain poorly modeled metal-binding sites. CheckMyMetal (CMM) is an easy-to-use metal-binding site validation server for macromolecules that is freely available at http://csgid.org/csgid/metal_sites. The CMM server can detect incorrect metal assignments as well as geometrical and other irregularities in the metal-binding sites. Guidelines for metal-site modeling and validation in macromolecules are illustrated by several practical examples grouped by the type of metal. These examples show CMM users (and crystallographers in general) problems they may encounter during the modeling of a specific metal ion.
Keywords: CheckMyMetal; coordination geometry; metal-binding environment; validation.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

