Transcriptome difference and potential crosstalk between liver and mammary tissue in mid-lactation primiparous dairy cows
- PMID: 28291785
- PMCID: PMC5349457
- DOI: 10.1371/journal.pone.0173082
Transcriptome difference and potential crosstalk between liver and mammary tissue in mid-lactation primiparous dairy cows
Abstract
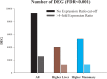
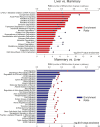
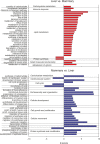
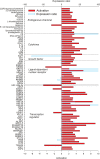
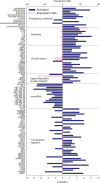
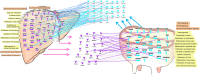
Liver and mammary gland are among the most important organs during lactation in dairy cows. With the purpose of understanding both the different and the complementary roles and the crosstalk of those two organs during lactation, a transcriptome analysis was performed on liver and mammary tissues of 10 primiparous dairy cows in mid-lactation. The analysis was performed using a 4×44K Bovine Agilent microarray chip. The transcriptome difference between the two tissues was analyzed using SAS JMP Genomics using ANOVA with a false discovery rate correction (FDR). The analysis uncovered >9,000 genes differentially expressed (DEG) between the two tissues with a FDR<0.001. The functional analysis of the DEG uncovered a larger metabolic (especially related to lipid) and inflammatory response capacity in liver compared with mammary tissue while the mammary tissue had a larger protein synthesis and secretion, proliferation/differentiation, signaling, and innate immune system capacity compared with the liver. A plethora of endogenous compounds, cytokines, and transcription factors were estimated to control the DEG between the two tissues. Compared with mammary tissue, the liver transcriptome appeared to be under control of a large array of ligand-dependent nuclear receptors and, among endogenous chemical, fatty acids and bacteria-derived compounds. Compared with liver, the transcriptome of the mammary tissue was potentially under control of a large number of growth factors and miRNA. The in silico crosstalk analysis between the two tissues revealed an overall large communication with a reciprocal control of lipid metabolism, innate immune system adaptation, and proliferation/differentiation. In summary the transcriptome analysis confirmed prior known differences between liver and mammary tissue, especially considering the indication of a larger metabolic activity in liver compared with the mammary tissue and the larger protein synthesis, communication, and proliferative capacity in mammary tissue compared with the liver. Relatively novel is the indication by the data that the transcriptome of the liver is highly regulated by dietary and bacteria-related compounds while the mammary transcriptome is more under control of hormones, growth factors, and miRNA. A large crosstalk between the two tissues with a reciprocal control of metabolism and innate immune-adaptation was indicated by the network analysis that allowed uncovering previously unknown crosstalk between liver and mammary tissue for several signaling molecules.
Conflict of interest statement
Figures

References
-
- Schingoethe DJ, Byers FM, Schelling GT (1988) Nutrient needs during critical periods of the life cycle In: Church D, editor. The Ruminant Animal: Digestive, Physiology, and Nutrition. Illinois, USA: Waveland Press Inc. pp. 421–447.
-
- Kuhn C, Freyer G, Weikard R, Goldammer T, Schwerin M (1999) Detection of QTL for milk production traits in cattle by application of a specifically developed marker map of BTA6. Anim Genet 30, 333–340. - PubMed
-
- Bergman EN, Brockman RP, Kaufman CF (1974) Glucose-Metabolism in Ruminants—Comparison of Whole-Body Turnover with Production by Gut, Liver, and Kidneys. Federation Proceedings 33, 1849–1854. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

