Systematic identification and characterization of long non-coding RNAs in mouse mature sperm
- PMID: 28291811
- PMCID: PMC5349675
- DOI: 10.1371/journal.pone.0173402
Systematic identification and characterization of long non-coding RNAs in mouse mature sperm
Abstract
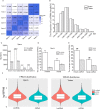
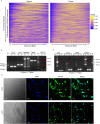
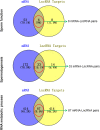
Increasing studies have shown that mature spermatozoa contain many transcripts including mRNAs and miRNAs. However, the expression profile of long non-coding RNAs (lncRNAs) in mammalian sperm has not been systematically investigated. Here, we used highly purified RNA to investigate lncRNA expression profiles in mouse mature sperm by stranded-specific RNA-seq. We identified 20,907 known and 4,088 novel lncRNAs transcripts, and the existence of intact lncRNAs was confirmed by RT-PCR and fluorescence in situ hybridization on two representative lncRNAs. Compared to round spermatids, 1,794 upregulated and 165 downregulated lncRNAs and 4,435 upregulated and 3,920 downregulated mRNAs were identified in sperm. Based on the "Cis and Trans" RNA-RNA interaction principle, we found 14,259 targeted coding genes of differently expressed lncRNAs. In terms of Gene ontology (GO) analysis, differentially expressed lncRNAs targeted genes mainly related to nucleic acid metabolic, protein modification, chromatin and histone modification, heterocycle compound metabolic, sperm function, spermatogenesis and other processes. In contrast, differentially expressed transcripts of mRNAs were highly enriched for protein metabolic process and RNA metabolic, spermatogenesis, sperm motility, cell cycle, chromatin organization, heterocycle and aromatic compound metabolic processes. Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis showed that the differentially expressed lncRNAs were involved in RNA transport, mRNA surveillance pathway, PI3K-Akt signaling pathway, AMPK signaling pathway, protein processing in endoplasmic reticulum. Metabolic pathways, mRNA surveillance pathway, AMPK signaling pathway, cell cycle, RNA transport splicesome and endocytosis incorporated with the differentially expressed mRNA. Furthermore, many lncRNAs were specifically expressed in testis/sperm, and 880 lncRNAs were conserved between human and mouse. In summary, this study provides a preliminary database valuable for identifying lncRNAs critical in the late stage of spermatogenesis or important for sperm function regulation, fertilization and early embryo development.
Conflict of interest statement
Figures



Similar articles
-
Profile and validation of dysregulated long non‑coding RNAs and mRNAs in ovarian cancer.Oncol Rep. 2018 Nov;40(5):2964-2976. doi: 10.3892/or.2018.6654. Epub 2018 Aug 17. Oncol Rep. 2018. PMID: 30132558
-
Systematic Analysis of Long Non-Coding RNAs and mRNAs in the Ovaries of Duroc Pigs During Different Follicular Stages Using RNA Sequencing.Int J Mol Sci. 2018 Jun 11;19(6):1722. doi: 10.3390/ijms19061722. Int J Mol Sci. 2018. PMID: 29891752 Free PMC article.
-
Expression profiles and characteristics of human lncRNA in normal and asthenozoospermia sperm†.Biol Reprod. 2019 Apr 1;100(4):982-993. doi: 10.1093/biolre/ioy253. Biol Reprod. 2019. PMID: 30517597
-
Long non-coding RNAs in human early embryonic development and their potential in ART.Hum Reprod Update. 2016 Dec;23(1):19-40. doi: 10.1093/humupd/dmw035. Epub 2016 Sep 21. Hum Reprod Update. 2016. PMID: 27655590 Review.
-
Long non-coding RNAs (lncRNAs) in spermatogenesis and male infertility.Reprod Biol Endocrinol. 2020 Oct 30;18(1):103. doi: 10.1186/s12958-020-00660-6. Reprod Biol Endocrinol. 2020. PMID: 33126901 Free PMC article. Review.
Cited by
-
Epigenetic transgenerational inheritance of testis pathology and Sertoli cell epimutations: generational origins of male infertility.Environ Epigenet. 2019 Aug 29;5(3):dvz013. doi: 10.1093/eep/dvz013. eCollection 2019 Jul. Environ Epigenet. 2019. PMID: 31528361 Free PMC article.
-
Maternal overnutrition programs hedonic and metabolic phenotypes across generations through sperm tsRNAs.Proc Natl Acad Sci U S A. 2019 May 21;116(21):10547-10556. doi: 10.1073/pnas.1820810116. Epub 2019 May 6. Proc Natl Acad Sci U S A. 2019. PMID: 31061112 Free PMC article.
-
Do Gametes Woo? Evidence for Their Nonrandom Union at Fertilization.Genetics. 2017 Oct;207(2):369-387. doi: 10.1534/genetics.117.300109. Genetics. 2017. PMID: 28978771 Free PMC article. Review.
-
Exploring the Characters of Non-Coding RNAs in Spermatogenesis and Male Infertility.Int J Mol Sci. 2025 Jan 28;26(3):1128. doi: 10.3390/ijms26031128. Int J Mol Sci. 2025. PMID: 39940895 Free PMC article. Review.
-
Dysregulation of lncRNA and circRNA Expression in Mouse Testes after Exposure to Triptolide.Curr Drug Metab. 2019;20(8):665-673. doi: 10.2174/1389200220666190729130020. Curr Drug Metab. 2019. PMID: 31362668 Free PMC article.
References
-
- Pessot CA, Brito M, Figueroa J, Concha II, Yanez A, Burzio LO. Presence of RNA in the sperm nucleus. Biochemical and biophysical research communications. 1989;158(1):272–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

