Fat2 and Lar Define a Basally Localized Planar Signaling System Controlling Collective Cell Migration
- PMID: 28292425
- PMCID: PMC5354100
- DOI: 10.1016/j.devcel.2017.02.003
Fat2 and Lar Define a Basally Localized Planar Signaling System Controlling Collective Cell Migration
Abstract
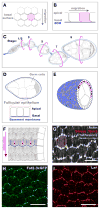
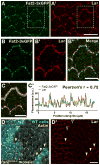
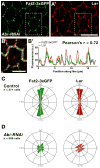
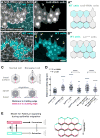
Collective migration of epithelial cells underlies diverse tissue-remodeling events, but the mechanisms that coordinate individual cell migratory behaviors for collective movement are largely unknown. Studying the Drosophila follicular epithelium, we show that the cadherin Fat2 and the receptor tyrosine phosphatase Lar function in a planar signaling system that coordinates leading and trailing edge dynamics between neighboring cells. Fat2 signals from each cell's trailing edge to induce leading edge protrusions in the cell behind, in part by stabilizing Lar's localization in these cells. Conversely, Lar signals from each cell's leading edge to stimulate trailing edge retraction in the cell ahead. Fat2/Lar signaling is similar to planar cell polarity signaling in terms of sub-cellular protein localization; however, Fat2/Lar signaling mediates short-range communication between neighboring cells instead of transmitting long-range information across a tissue. This work defines a key mechanism promoting epithelial migration and establishes a different paradigm for planar cell-cell signaling.
Keywords: Drosophila; Fat2; Lar; cadherin; collective cell migration; egg chamber; epithelium.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures



Comment in
-
Fat2 and Lar Dance a Pas de Deux during Collective Cell Migration.Dev Cell. 2017 Mar 13;40(5):425-426. doi: 10.1016/j.devcel.2017.02.025. Dev Cell. 2017. PMID: 28292420
References
-
- Aigouy B, Farhadifar R, Staple DB, Sagner A, Röper JC, Jülicher F, Eaton S. Cell Flow Reorients the Axis of Planar Polarity in the Wing Epithelium of Drosophila. Cell. 2010;142:773–786. - PubMed
-
- Aurich F, Dahmann C. A Mutation in fat2 Uncouples Tissue Elongation from Global Tissue Rotation. CellReports. 2016;14:2503–2510. - PubMed
-
- Bateman J, Reddy RS, Saito H, Van Vactor D. The receptor tyrosine phosphatase Dlar and integrins organize actin filaments in the Drosophila follicular epithelium. Current Biology. 2001;11:1317–1327. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

