Phenylalanine isotope pulse method to measure effect of sepsis on protein breakdown and membrane transport in the pig
- PMID: 28292760
- PMCID: PMC5494580
- DOI: 10.1152/ajpendo.00351.2016
Phenylalanine isotope pulse method to measure effect of sepsis on protein breakdown and membrane transport in the pig
Abstract
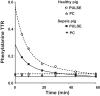
The primed-continuous (PC) phenylalanine (Phe) stable isotope infusion methodology is often used as a proxy for measuring whole body protein breakdown (WbPB) in sepsis. It is unclear if WbPB data obtained by an easy-to-use single IV Phe isotope pulse administration (PULSE) are comparable to those by PC. Compartmental modeling with PULSE could provide us more insight in WbPB in sepsis. Therefore, in the present study, we compared PULSE with PC as proxy for WbPB in an instrumented pig model with Pseudomonas aeruginosa-induced severe sepsis (Healthy: n = 9; Sepsis: n = 13). Seventeen hours after sepsis induction, we compared the Wb rate of appearance (WbRa) of Phe obtained by PC (L-[ring-13C6]Phe) and PULSE (L-[15N]Phe) in arterial plasma using LC-MS/MS and (non)compartmental modeling. PULSE-WbRa was highly correlated with PC-WbRa (r = 0.732, P < 0.0001) and WbPB (r = 0.897, P < 0.0001) independent of the septic state. PULSE-WbRa was 1.6 times higher than PC-WbRa (P < 0.001). Compartmental and noncompartmental PULSE modeling provide comparable WbRa values, although compartmental modeling was more sensitive. WbPB was elevated in sepsis (Healthy: 3,378 ± 103; Sepsis: 4,333 ± 160 nmol·kg BW-1·min-1, P = 0.0002). With PULSE, sepsis was characterized by an increase of the metabolic shunting (Healthy: 3,021 ± 347; Sepsis: 4,233 ± 344 nmol·kg BW-1·min-1, P = 0.026). Membrane transport capacity was the same. Both PC and PULSE methods are able to assess changes in WbRa of plasma Phe reflecting WbPB changes with high sensitivity, independent of the (patho)physiological state. The easy-to-use (non)compartmental PULSE reflects better the real WbPB than PC. With PULSE compartmental analysis, we conclude that the membrane transport capacity for amino acids is not compromised in severe sepsis.
Keywords: phenylalanine; pig; protein breakdown; pulse stable isotope method; sepsis.
Copyright © 2017 the American Physiological Society.
Figures







References
-
- Bregendahl K, Yang X, Liu L, Yen JT, Rideout TC, Shen Y, Werchola G, Fan MZ. Fractional protein synthesis rates are similar when measured by intraperitoneal or intravenous flooding doses of L-[ring-2H5]phenylalanine in combination with a rapid regimen of sampling in piglets. J Nutr 138: 1976–1981, 2008. - PubMed
-
- Bruins MJ, Soeters PB, Deutz NE. Endotoxemia affects organ protein metabolism differently during prolonged feeding in pigs. J Nutr 130: 3003–3013, 2000. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

