Fission yeast myosin I facilitates PI(4,5)P2-mediated anchoring of cytoplasmic dynein to the cortex
- PMID: 28292899
- PMCID: PMC5380020
- DOI: 10.1073/pnas.1615883114
Fission yeast myosin I facilitates PI(4,5)P2-mediated anchoring of cytoplasmic dynein to the cortex
Abstract
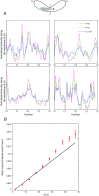
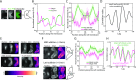
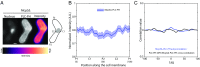
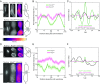
Several key processes in the cell, such as vesicle transport and spindle positioning, are mediated by the motor protein cytoplasmic dynein, which produces force on the microtubule. For the functions that require movement of the centrosome and the associated nuclear material, dynein needs to have a stable attachment at the cell cortex. In fission yeast, Mcp5 is the anchor protein of dynein and is required for the oscillations of the horsetail nucleus during meiotic prophase. Although the role of Mcp5 in anchoring dynein to the cortex has been identified, it is unknown how Mcp5 associates with the membrane as well as the importance of the underlying attachment to the nuclear oscillations. Here, we set out to quantify Mcp5 organization and identify the binding partner of Mcp5 at the membrane. We used confocal and total internal reflection fluorescence microscopy to count the number of Mcp5 foci and the number of Mcp5 molecules in an individual focus. Further, we quantified the localization pattern of Mcp5 in fission yeast zygotes and show by perturbation of phosphatidylinositol 4-phosphate 5-kinase that Mcp5 binds to phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2]. Remarkably, we discovered that the myosin I protein in fission yeast, Myo1, which is required for organization of sterol-rich domains in the cell membrane, facilitates the localization of Mcp5 and that of cytoplasmic dynein on the membrane. Finally, we demonstrate that Myo1-facilitated association of Mcp5 and dynein to the membrane determines the dynamics of nuclear oscillations and, in essence, dynein activity.
Keywords: Mcp5; PIP2; cortical anchoring; cytoplasmic dynein; myosin I.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


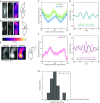
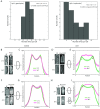

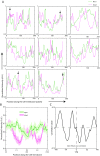

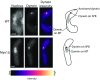

References
-
- Ding DQ, Chikashige Y, Haraguchi T, Hiraoka Y. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J Cell Sci. 1998;111(Pt 6):701–712. - PubMed
-
- Chikashige Y, et al. Telomere-led premeiotic chromosome movement in fission yeast. Science. 1994;264(5156):270–273. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

