Histone peptide microarray screen of chromo and Tudor domains defines new histone lysine methylation interactions
- PMID: 28293301
- PMCID: PMC5348760
- DOI: 10.1186/s13072-017-0117-5
Histone peptide microarray screen of chromo and Tudor domains defines new histone lysine methylation interactions
Abstract
Background: Histone posttranslational modifications (PTMs) function to regulate chromatin structure and function in part through the recruitment of effector proteins that harbor specialized "reader" domains. Despite efforts to elucidate reader domain-PTM interactions, the influence of neighboring PTMs and the target specificity of many reader domains is still unclear. The aim of this study was to use a high-throughput histone peptide microarray platform to interrogate 83 known and putative histone reader domains from the chromo and Tudor domain families to identify their interactions and characterize the influence of neighboring PTMs on these interactions.
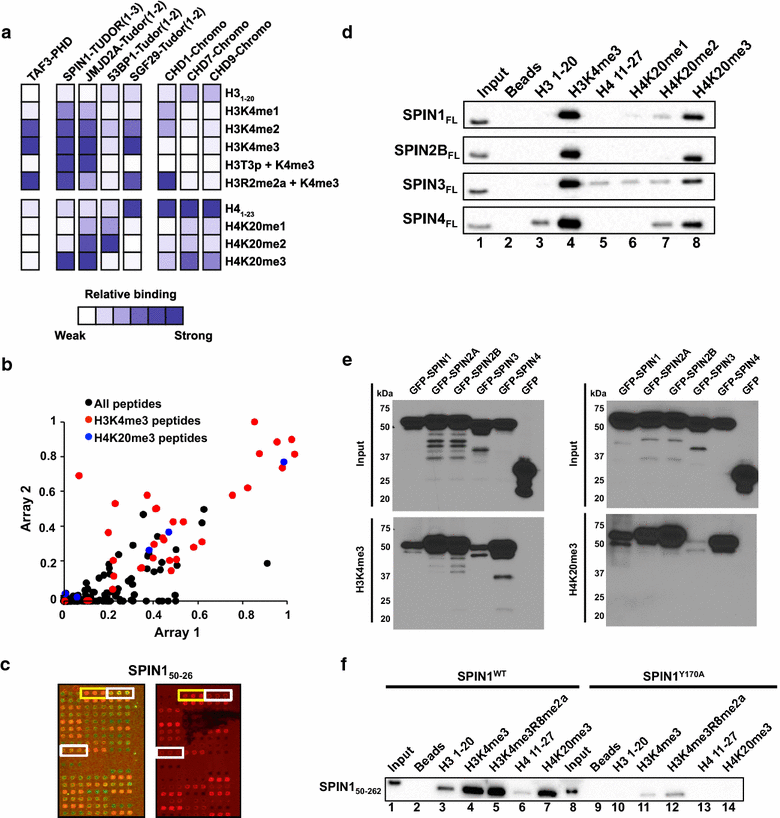
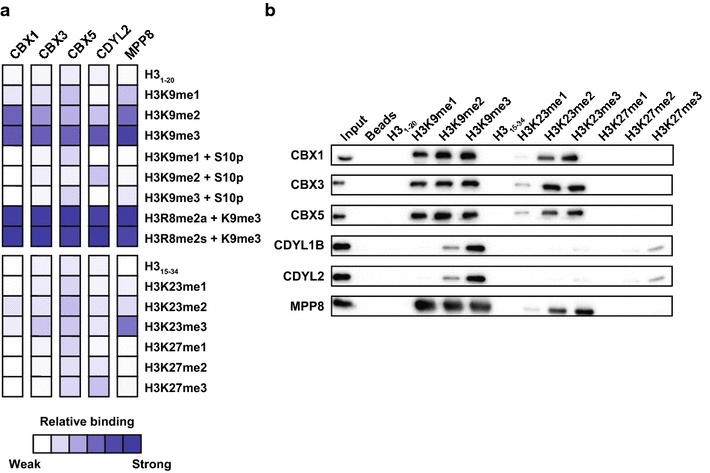
Results: Nearly a quarter of the chromo and Tudor domains screened showed interactions with histone PTMs by peptide microarray, revealing known and several novel methyllysine interactions. Specifically, we found that the CBX/HP1 chromodomains that recognize H3K9me also recognize H3K23me2/3-a poorly understood histone PTM. We also observed that, in addition to their interaction with H3K4me3, Tudor domains of the Spindlin family also recognized H4K20me3-a previously uncharacterized interaction. Several Tudor domains also showed novel interactions with H3K4me as well.
Conclusions: These results provide an important resource for the epigenetics and chromatin community on the interactions of many human chromo and Tudor domains. They also provide the basis for additional studies into the functional significance of the novel interactions that were discovered.
Keywords: Chromatin; Chromodomain; Histone methylation; Peptide microarray; Tudor domain.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

