Measurement of DNA Length Changes Upon CpG Hypermethylation by Microfluidic Molecular Stretching
- PMID: 28293464
- PMCID: PMC5225679
- DOI: 10.3727/215517916X693087
Measurement of DNA Length Changes Upon CpG Hypermethylation by Microfluidic Molecular Stretching
Abstract
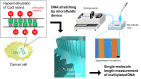
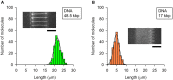
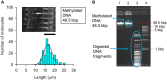
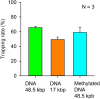
Abnormal DNA methylation in CpG-rich promoters is recognized as a distinct molecular feature of precursor lesions to cancer. Such unintended methylation can occur during in vitro differentiation of stem cells. It takes place in a subset of genes during the differentiation or expansion of stem cell derivatives under general culture conditions, which may need to be monitored in future cell transplantation studies. Here we demonstrate a microfluidic device for investigating morphological length changes in DNA methylation. Arrayed polymer chains of single DNA molecules were fluorescently observed by parallel trapping and stretching in the microfluidic channel. This observational study revealed that the shortened DNA length is due to the increased rigidity of the methylated DNA molecule. The trapping rate of the device for DNA molecules was substantially unaffected by changes in the CpG methylation.
Keywords: Cytosine-guanine dinucleotides (CpG); DNA methylation; Microfluidic device; Single-molecule detection.
Figures

References
-
- Brena RM, Huang TH, Plass C. Toward a human epigenome. Nat Genet. 2006;38:1359–60. - PubMed
-
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–54. - PubMed
-
- Esteller M. CpG island hypermethylation and tumor suppressor genes: A booming present, a brighter future. Oncogene 2002;21:5427–40. - PubMed
-
- Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer 2004;4:988–93. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

