Integrating miRNA and mRNA Expression Profiling Uncovers miRNAs Underlying Fat Deposition in Sheep
- PMID: 28293627
- PMCID: PMC5331317
- DOI: 10.1155/2017/1857580
Integrating miRNA and mRNA Expression Profiling Uncovers miRNAs Underlying Fat Deposition in Sheep
Abstract
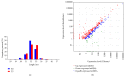
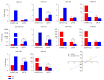
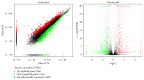
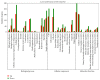
MicroRNAs (miRNAs) are endogenous, noncoding RNAs that regulate various biological processes including adipogenesis and fat metabolism. Here, we adopted a deep sequencing approach to determine the identity and abundance of miRNAs involved in fat deposition in adipose tissues from fat-tailed (Kazakhstan sheep, KS) and thin-tailed (Tibetan sheep, TS) sheep breeds. By comparing HiSeq data of these two breeds, 539 miRNAs were shared in both breeds, whereas 179 and 97 miRNAs were uniquely expressed in KS and TS, respectively. We also identified 35 miRNAs that are considered to be putative novel miRNAs. The integration of miRNA-mRNA analysis revealed that miRNA-associated targets were mainly involved in the gene ontology (GO) biological processes concerning cellular process and metabolic process, and miRNAs play critical roles in fat deposition through their ability to regulate fundamental pathways. These pathways included the MAPK signaling pathway, FoxO and Wnt signaling pathway, and focal adhesion. Taken together, our results define miRNA expression signatures that may contribute to fat deposition and lipid metabolism in sheep.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Genome-wide analysis of microRNAs identifies the lipid metabolism pathway to be a defining factor in adipose tissue from different sheep.Sci Rep. 2015 Dec 22;5:18470. doi: 10.1038/srep18470. Sci Rep. 2015. PMID: 26690086 Free PMC article.
-
Transcriptome reveals key microRNAs involved in fat deposition between different tail sheep breeds.PLoS One. 2022 Mar 1;17(3):e0264804. doi: 10.1371/journal.pone.0264804. eCollection 2022. PLoS One. 2022. PMID: 35231067 Free PMC article.
-
A comprehensive microRNA expression profile of the backfat tissue from castrated and intact full-sib pair male pigs.BMC Genomics. 2014 Jan 20;15:47. doi: 10.1186/1471-2164-15-47. BMC Genomics. 2014. PMID: 24443800 Free PMC article.
-
The physiological and pathophysiological roles of adipocyte miRNAs.Biochem Cell Biol. 2013 Aug;91(4):195-202. doi: 10.1139/bcb-2012-0053. Epub 2013 Jan 25. Biochem Cell Biol. 2013. PMID: 23859012 Review.
-
MicroRNA regulatory networks in human adipose tissue and obesity.Nat Rev Endocrinol. 2015 May;11(5):276-88. doi: 10.1038/nrendo.2015.25. Epub 2015 Mar 3. Nat Rev Endocrinol. 2015. PMID: 25732520 Review.
Cited by
-
Identification and characterization of microRNAs in the intestinal tissues of sheep (Ovis aries).PLoS One. 2018 Feb 28;13(2):e0193371. doi: 10.1371/journal.pone.0193371. eCollection 2018. PLoS One. 2018. PMID: 29489866 Free PMC article.
-
Integrated Metabolomics and Transcriptomics Analyses Reveal the Candidate Genes Regulating the Meat Quality Change by Castration in Yudong Black Goats (Capra hircus).Genes (Basel). 2023 Dec 27;15(1):43. doi: 10.3390/genes15010043. Genes (Basel). 2023. PMID: 38254933 Free PMC article.
-
MiRNA-Seq reveals key MicroRNAs involved in fat metabolism of sheep liver.Front Genet. 2023 Mar 9;14:985764. doi: 10.3389/fgene.2023.985764. eCollection 2023. Front Genet. 2023. PMID: 36968587 Free PMC article.
-
Comprehensive Gene Expression Profiling Analysis of Adipose Tissue in Male Individuals from Fat- and Thin-Tailed Sheep Breeds.Animals (Basel). 2023 Nov 10;13(22):3475. doi: 10.3390/ani13223475. Animals (Basel). 2023. PMID: 38003093 Free PMC article.
-
Genetics of the phenotypic evolution in sheep: a molecular look at diversity-driving genes.Genet Sel Evol. 2022 Sep 9;54(1):61. doi: 10.1186/s12711-022-00753-3. Genet Sel Evol. 2022. PMID: 36085023 Free PMC article. Review.
References
-
- Davidson A. Oxford Companion to Food. Oxford, UK: Oxford University Press; 1999.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

