Proteome and Protein Network Analyses of Memory T Cells Find Altered Translation and Cell Stress Signaling in Treated Human Immunodeficiency Virus Patients Exhibiting Poor CD4 Recovery
- PMID: 28293663
- PMCID: PMC4866573
- DOI: 10.1093/ofid/ofw037
Proteome and Protein Network Analyses of Memory T Cells Find Altered Translation and Cell Stress Signaling in Treated Human Immunodeficiency Virus Patients Exhibiting Poor CD4 Recovery
Abstract
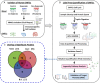
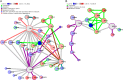
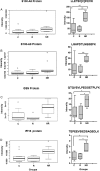
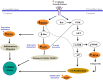
Background. Human immunodeficiency virus (HIV) patients who experience poor CD4 T-cell recovery despite viral suppression during antiretroviral therapy (ART) are known as immunological nonresponders. The molecular mechanism(s) underlying incomplete immune restoration during ART is not fully understood. Methods. Label-free quantitative proteomics on single-cell type central memory T cells were used to reveal relative protein abundance changes between nonresponder, responder (good CD4 recovery during ART), and healthy individuals. Proteome changes were analyzed by protein pathway and network analyses and verified by selected reaction monitoring mass spectrometry. Results. Proteomic analysis across groups detected 155 significant proteins from 1500 nonredundant proteins. Pathway and network analyses revealed dysregulation in mammalian target of rapamycin and protein translation-related proteins and decreases in stress response-related proteins for nonresponder subjects compared with responders and controls. Actin cytoskeleton signaling was increased for HIV responders and nonresponders alike. Conclusions. Memory T cells from immunologic nonresponders have increases in proteins related to motility and protein translation and decreases in proteins capable of responding to cellular stresses compared with responders and controls. The potential for T cells to manage stress and modulate metabolism may contribute to their capacity to reconstitute a lymphopenic host.
Keywords: ART; CMTC; HIV; SRM; proteomics.
Figures

References
-
- Hammer SM, Squires KE, Hughes MD et al. . A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N Engl J Med 1997; 337:725–33. - PubMed
-
- Gulick RM, Mellors JW, Havlir D et al. . Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med 1997; 337:734–9. - PubMed
-
- Powderly WG, Landay A, Lederman MM. Recovery of the immune system with antiretroviral therapy: the end of opportunism? JAMA 1998; 280:72–7. - PubMed
-
- Lewden C, Chene G, Morlat P et al. . HIV-infected adults with a CD4 cell count greater than 500 cells/mm3 on long-term combination antiretroviral therapy reach same mortality rates as the general population. J Acquir Immune Defic Syndr 2007; 46:72–7. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

