Engineered Split-TET2 Enzyme for Inducible Epigenetic Remodeling
- PMID: 28294608
- PMCID: PMC5385525
- DOI: 10.1021/jacs.7b01459
Engineered Split-TET2 Enzyme for Inducible Epigenetic Remodeling
Abstract
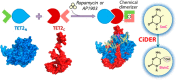
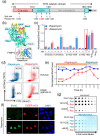
The Ten-eleven translocation (TET) family of 5-methylcytosine (5mC) dioxygenases catalyze the conversion of 5mC into 5-hydroxymethylcytosine (5hmC) and further oxidized species to promote active DNA demethylation. Here we engineered a split-TET2 enzyme to enable temporal control of 5mC oxidation and subsequent remodeling of epigenetic states in mammalian cells. We further demonstrate the use of this chemically inducible system to dissect the correlation between DNA hydroxymethylation and chromatin accessibility in the mammalian genome. This chemical-inducible epigenome remodeling tool will find broad use in interrogating cellular systems without altering the genetic code, as well as in probing the epigenotype-phenotype relations in various biological systems.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Spruijt C. G.; Gnerlich F.; Smits A. H.; Pfaffeneder T.; Jansen P. W.; Bauer C.; Munzel M.; Wagner M.; Muller M.; Khan F.; Eberl H. C.; Mensinga A.; Brinkman A. B.; Lephikov K.; Muller U.; Walter J.; Boelens R.; van Ingen H.; Leonhardt H.; Carell T.; Vermeulen M. Cell 2013, 152, 1146–59. 10.1016/j.cell.2013.02.004. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

