Reverse Transcription in the Saccharomyces cerevisiae Long-Terminal Repeat Retrotransposon Ty3
- PMID: 28294975
- PMCID: PMC5371799
- DOI: 10.3390/v9030044
Reverse Transcription in the Saccharomyces cerevisiae Long-Terminal Repeat Retrotransposon Ty3
Abstract
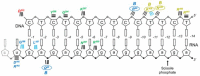
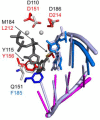
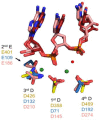
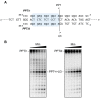
Converting the single-stranded retroviral RNA into integration-competent double-stranded DNA is achieved through a multi-step process mediated by the virus-coded reverse transcriptase (RT). With the exception that it is restricted to an intracellular life cycle, replication of the Saccharomyces cerevisiae long terminal repeat (LTR)-retrotransposon Ty3 genome is guided by equivalent events that, while generally similar, show many unique and subtle differences relative to the retroviral counterparts. Until only recently, our knowledge of RT structure and function was guided by a vast body of literature on the human immunodeficiency virus (HIV) enzyme. Although the recently-solved structure of Ty3 RT in the presence of an RNA/DNA hybrid adds little in terms of novelty to the mechanistic basis underlying DNA polymerase and ribonuclease H activity, it highlights quite remarkable topological differences between retroviral and LTR-retrotransposon RTs. The theme of overall similarity but distinct differences extends to the priming mechanisms used by Ty3 RT to initiate (-) and (+) strand DNA synthesis. The unique structural organization of the retrotransposon enzyme and interaction with its nucleic acid substrates, with emphasis on polypurine tract (PPT)-primed initiation of (+) strand synthesis, is the subject of this review.
Keywords: DNA polymerase; Ty3; retroelement; retrotransposon; reverse transcriptase; reverse transcription; ribonuclease H (RNase H).
Conflict of interest statement
The authors declare no conflict of interest. The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures



References
-
- Telesnitsky A., Goff S.P. Reverse transcriptase and the generation of retroviral DNA. In: Coffin J.M., Hughes S.H., Varmus H.E., editors. Retroviruses. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 1997. pp. 121–160. - PubMed
-
- Nowak E., Potrzebowski W., Konarev P.V., Rausch J.W., Bona M.K., Svergun D.I., Bujnicki J.M., Le Grice S.F., Nowotny M. Structural analysis of monomeric retroviral reverse transcriptase in complex with an RNA/DNA hybrid. Nucleic Acids Res. 2013;41:3874–3887. doi: 10.1093/nar/gkt053. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

