RBM10-TFE3 Renal Cell Carcinoma: A Potential Diagnostic Pitfall Due to Cryptic Intrachromosomal Xp11.2 Inversion Resulting in False-negative TFE3 FISH
- PMID: 28296677
- PMCID: PMC5391276
- DOI: 10.1097/PAS.0000000000000835
RBM10-TFE3 Renal Cell Carcinoma: A Potential Diagnostic Pitfall Due to Cryptic Intrachromosomal Xp11.2 Inversion Resulting in False-negative TFE3 FISH
Abstract
Xp11 translocation renal cell carcinoma (RCC) are defined by chromosome translocations involving the Xp11 breakpoint which results in one of a variety of TFE3 gene fusions. TFE3 break-apart florescence in situ hybridization (FISH) assays are generally preferred to TFE3 immunohistochemistry (IHC) as a means of confirming the diagnosis in archival material, as FISH is less sensitive to the variable fixation which can result in false positive or false negative IHC. Prompted by a case report in the cytogenetics literature, we identify 3 cases of Xp11 translocation RCC characterized by a subtle chromosomal inversion involving the short arm of the X chromosome, resulting in an RBM10-TFE3 gene fusion. TFE3 rearrangement was not detected by conventional TFE3 break-apart FISH, but was suggested by strong diffuse TFE3 immunoreactivity in a clean background. We then developed novel fosmid probes to detect the RBM10-TFE3 gene fusion in archival material. These cases validate RBM10-TFE3 as a recurrent gene fusion in Xp11 translocation RCC, illustrate a source of false-negative TFE3 break-apart FISH, and highlight the complementary role of TFE3 IHC and TFE3 FISH.
Figures

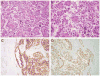
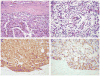


References
-
- Argani P, Antonescu CR, Illei PB, Lui MY, Timmons CF, Newbury R, Reuter VE, Garvin AJ, Perez-Atayde AR, Fletcher JA, Beckwith JB, Bridge JA, Ladanyi M. Primary renal neoplasms with the ASPL-TFE3 gene fusion of alveolar soft part sarcoma: A distinctive tumor entity previously included among renal cell carcinomas of children and adolescents. Am J Pathol. 2001;159:179–92. - PMC - PubMed
-
- Argani P, Antonescu CR, Couturier J, Fournet J, Sciot R, Debiec-Rychter M, Hutchinson B, Reuter VE, Boccon-Gibod L, Timmons C, Hafez N, Ladanyi M. PRCC-TFE3 renal carcinomas: Morphologic, immunohistochemical, ultrastructural, and molecular analysis of an entity associated with the t(X;1)(p11. 2;q21) Am J Surg Pathol. 2002;26:1553–66. - PubMed
-
- Clark J, Lu YJ, Sidhar SK, Parker C, Gill S, Smedley D, Hamoudi R, Linehan WM, Shipley J, Cooper CS. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene. 1997;15:2233–9. - PubMed
-
- Argani P, Lui MY, Couturier J, Bouvier R, Fournet J, Ladanyi M. A novel CLTC-TFE3 gene fusion in pediatric renal adenocarcinoma with t(X;17)(p11. 2;q23) Oncogene. 2003;22:5374–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

