Whole-exome sequencing identifies recurrent SF3B1 R625 mutation and comutation of NF1 and KIT in mucosal melanoma
- PMID: 28296713
- PMCID: PMC5470740
- DOI: 10.1097/CMR.0000000000000345
Whole-exome sequencing identifies recurrent SF3B1 R625 mutation and comutation of NF1 and KIT in mucosal melanoma
Abstract
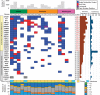
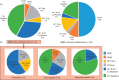
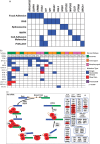
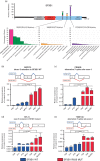
Mucosal melanomas are a rare subtype of melanoma, arising in mucosal tissues, which have a very poor prognosis due to the lack of effective targeted therapies. This study aimed to better understand the molecular landscape of these cancers and find potential new therapeutic targets. Whole-exome sequencing was performed on mucosal melanomas from 19 patients and 135 sun-exposed cutaneous melanomas, with matched peripheral blood samples when available. Mutational profiles were compared between mucosal subgroups and sun-exposed cutaneous melanomas. Comparisons of molecular profiles identified 161 genes enriched in mucosal melanoma (P<0.05). KIT and NF1 were frequently comutated (32%) in the mucosal subgroup, with a significantly higher incidence than that in cutaneous melanoma (4%). Recurrent SF3B1 R625H/S/C mutations were identified and validated in 7 of 19 (37%) mucosal melanoma patients. Mutations in the spliceosome pathway were found to be enriched in mucosal melanomas when compared with cutaneous melanomas. Alternative splicing in four genes were observed in SF3B1-mutant samples compared with the wild-type samples. This study identified potential new therapeutic targets for mucosal melanoma, including comutation of NF1 and KIT, and recurrent R625 mutations in SF3B1. This is the first report of SF3B1 R625 mutations in vulvovaginal mucosal melanoma, with the largest whole-exome sequencing project of mucosal melanomas to date. The results here also indicated that the mutations in SF3B1 lead to alternative splicing in multiple genes. These findings expand our knowledge of this rare disease.
Figures
References
-
- Furney SJ, Turajlic S, Stamp G, Nohadani M, Carlisle A, Thomas JM, et al. Genome sequencing of mucosal melanomas reveals that they are driven by distinct mechanisms from cutaneous melanoma. J Pathol 2013; 230:261–269. - PubMed
-
- Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med 2005; 353:2135–2147. - PubMed
-
- Del Vecchio M, Di Guardo L, Ascierto PA, Grimaldi AM, Sileni VC, Pigozzo J, et al. Efficacy and safety of ipilimumab 3 mg/kg in patients with pretreated, metastatic, mucosal melanoma. Eur J Cancer 2014; 50:121–127. - PubMed
-
- Spencer KR, Mehnert JM. Mucosal melanoma: epidemiology, biology and treatment. Cancer Treat Res 2016; 167:295–320. - PubMed
-
- Postow MA, Hamid O, Carvajal RD. Mucosal melanoma: pathogenesis, clinical behavior, and management. Curr Oncol Rep 2012; 14:441–448. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

