Transcriptomic responses to environmental temperature by turtles with temperature-dependent and genotypic sex determination assessed by RNAseq inform the genetic architecture of embryonic gonadal development
- PMID: 28296881
- PMCID: PMC5352168
- DOI: 10.1371/journal.pone.0172044
Transcriptomic responses to environmental temperature by turtles with temperature-dependent and genotypic sex determination assessed by RNAseq inform the genetic architecture of embryonic gonadal development
Abstract
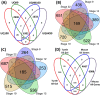
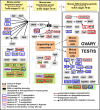
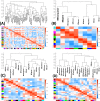
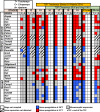
Vertebrate sexual fate is decided primarily by the individual's genotype (GSD), by the environmental temperature during development (TSD), or both. Turtles exhibit TSD and GSD, making them ideal to study the evolution of sex determination. Here we analyze temperature-specific gonadal transcriptomes (RNA-sequencing validated by qPCR) of painted turtles (Chrysemys picta TSD) before and during the thermosensitive period, and at equivalent stages in soft-shell turtles (Apalone spinifera-GSD), to test whether TSD's and GSD's transcriptional circuitry is identical but deployed differently between mechanisms. Our data show that most elements of the mammalian urogenital network are active during turtle gonadogenesis, but their transcription is generally more thermoresponsive in TSD than GSD, and concordant with their sex-specific function in mammals [e.g., upregulation of Amh, Ar, Esr1, Fog2, Gata4, Igf1r, Insr, and Lhx9 at male-producing temperature, and of β-catenin, Foxl2, Aromatase (Cyp19a1), Fst, Nf-kb, Crabp2 at female-producing temperature in Chrysemys]. Notably, antagonistic elements in gonadogenesis (e.g., β-catenin and Insr) were thermosensitive only in TSD early-embryos. Cirbp showed warm-temperature upregulation in both turtles disputing its purported key TSD role. Genes that may convert thermal inputs into sex-specific development (e.g., signaling and hormonal pathways, RNA-binding and heat-shock) were differentially regulated. Jak-Stat, Nf-κB, retinoic-acid, Wnt, and Mapk-signaling (not Akt and Ras-signaling) potentially mediate TSD thermosensitivity. Numerous species-specific ncRNAs (including Xist) were differentially-expressed, mostly upregulated at colder temperatures, as were unannotated loci that constitute novel TSD candidates. Cirbp showed warm-temperature upregulation in both turtles. Consistent transcription between turtles and alligator revealed putatively-critical reptilian TSD elements for male (Sf1, Amh, Amhr2) and female (Crabp2 and Hspb1) gonadogenesis. In conclusion, while preliminary, our data helps illuminate the regulation and evolution of vertebrate sex determination, and contribute genomic resources to guide further research into this fundamental biological process.
Conflict of interest statement
Figures


References
-
- Valenzuela N, Lance VA, eds. Temperature dependent sex determination in vertebrates. Washington, DC: Smithsonian Books; 2004.
-
- Bull JJ. Evolution of sex determining mechanisms. 1983:173–84.
-
- Deeming DC, Ferguson MWJ. Physiological effects of incubation temperature on embryonic development in reptiles and birds In: Deeming DC, Ferguson MWJ, editors. 2004: Cambridge University Press; 1991. p. 147–71.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

