Clinical Implications of Genetic Mutations in Myelodysplastic Syndrome
- PMID: 28297619
- PMCID: PMC5455680
- DOI: 10.1200/JCO.2016.71.0806
Clinical Implications of Genetic Mutations in Myelodysplastic Syndrome
Abstract
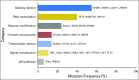
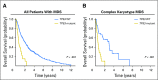
Myelodysplastic syndrome (MDS) is clonal disorder characterized by ineffective hematopoiesis and a tendency to evolve into acute myeloid leukemia (AML). Genetic studies have enabled the identification of a set of recurrently mutated genes central to the pathogenesis of MDS, which can be organized into a limited number of cellular processes, including RNA splicing, epigenetic and traditional transcriptional regulation, and signal transduction. The sequential accumulation of mutations drives disease evolution from asymptomatic clonal hematopoiesis to frank MDS, and, ultimately, to secondary AML. This detailed understanding of the molecular landscape of MDS, coupled with the emergence of cost- and time-effective methodologies for DNA sequencing has led to the introduction of genetic studies into the clinical realm. Here, we review recent advances in our genetic understanding of MDS, with a particular focus on the emerging role for mutational data in clinical management as a potential tool to assist in diagnosis, risk stratification, and therapeutic decision-making.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

