m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells
- PMID: 28297667
- PMCID: PMC5479356
- DOI: 10.1016/j.celrep.2017.02.059
m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells
Abstract
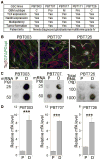
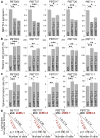
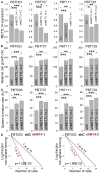
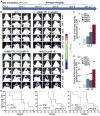
RNA modifications play critical roles in important biological processes. However, the functions of N6-methyladenosine (m6A) mRNA modification in cancer biology and cancer stem cells remain largely unknown. Here, we show that m6A mRNA modification is critical for glioblastoma stem cell (GSC) self-renewal and tumorigenesis. Knockdown of METTL3 or METTL14, key components of the RNA methyltransferase complex, dramatically promotes human GSC growth, self-renewal, and tumorigenesis. In contrast, overexpression of METTL3 or inhibition of the RNA demethylase FTO suppresses GSC growth and self-renewal. Moreover, inhibition of FTO suppresses tumor progression and prolongs lifespan of GSC-grafted mice substantially. m6A sequencing reveals that knockdown of METTL3 or METTL14 induced changes in mRNA m6A enrichment and altered mRNA expression of genes (e.g., ADAM19) with critical biological functions in GSCs. In summary, this study identifies the m6A mRNA methylation machinery as promising therapeutic targets for glioblastoma.
Keywords: FTO inhibitor; METTL14; METTL3; glioblastoma stem cells; m(6)A modification.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Allegra A, Alonci A, Penna G, Innao V, Gerace D, Rotondo F, Musolino C. The cancer stem cell hypothesis: a guide to potential molecular targets. Cancer Invest. 2014;32:470–495. - PubMed
-
- Anido J, Sáez-Borderías A, Gonzàlez-Juncà A, Rodón L, Folch G, Carmona MA, Prieto-Sánchez RM, Barba I, Martínez-Sáez E, Prudkin L, et al. TGF-β receptor inhibitors target the CD44(high)/Id1(high) glioma-initiating cell population in human glioblastoma. Cancer Cell. 2010;18:655–668. - PubMed
-
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. - PubMed
-
- Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem. 1994;269:17697–17704. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

