Bacterial protein meta-interactomes predict cross-species interactions and protein function
- PMID: 28298180
- PMCID: PMC5353844
- DOI: 10.1186/s12859-017-1585-0
Bacterial protein meta-interactomes predict cross-species interactions and protein function
Abstract
Background: Protein-protein interactions (PPIs) can offer compelling evidence for protein function, especially when viewed in the context of proteome-wide interactomes. Bacteria have been popular subjects of interactome studies: more than six different bacterial species have been the subjects of comprehensive interactome studies while several more have had substantial segments of their proteomes screened for interactions. The protein interactomes of several bacterial species have been completed, including several from prominent human pathogens. The availability of interactome data has brought challenges, as these large data sets are difficult to compare across species, limiting their usefulness for broad studies of microbial genetics and evolution.
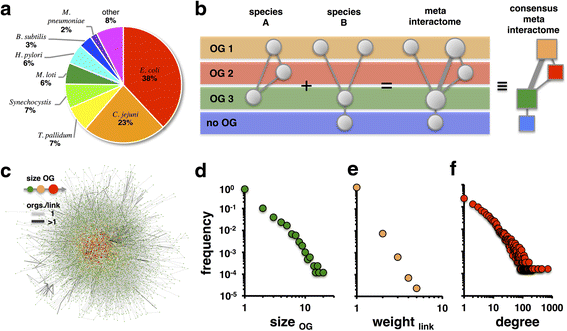
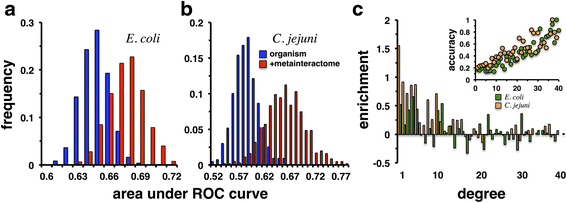
Results: In this study, we use more than 52,000 unique protein-protein interactions (PPIs) across 349 different bacterial species and strains to determine their conservation across data sets and taxonomic groups. When proteins are collapsed into orthologous groups (OGs) the resulting meta-interactome still includes more than 43,000 interactions, about 14,000 of which involve proteins of unknown function. While conserved interactions provide support for protein function in their respective species data, we found only 429 PPIs (~1% of the available data) conserved in two or more species, rendering any cross-species interactome comparison immediately useful. The meta-interactome serves as a model for predicting interactions, protein functions, and even full interactome sizes for species with limited to no experimentally observed PPI, including Bacillus subtilis and Salmonella enterica which are predicted to have up to 18,000 and 31,000 PPIs, respectively.
Conclusions: In the course of this work, we have assembled cross-species interactome comparisons that will allow interactomics researchers to anticipate the structures of yet-unexplored microbial interactomes and to focus on well-conserved yet uncharacterized interactors for further study. Such conserved interactions should provide evidence for important but yet-uncharacterized aspects of bacterial physiology and may provide targets for anti-microbial therapies.
Keywords: Genome evolution; Interactome; Networks; Protein interactions.
Figures





Similar articles
-
Mycobacterium tuberculosis and Clostridium difficille interactomes: demonstration of rapid development of computational system for bacterial interactome prediction.Microb Inform Exp. 2012 Mar 21;2:4. doi: 10.1186/2042-5783-2-4. Microb Inform Exp. 2012. PMID: 22587966 Free PMC article.
-
Predicting the interactome of Xanthomonas oryzae pathovar oryzae for target selection and DB service.BMC Bioinformatics. 2008 Jan 24;9:41. doi: 10.1186/1471-2105-9-41. BMC Bioinformatics. 2008. PMID: 18215330 Free PMC article.
-
Interactome evolution: insights from genome-wide analyses of protein-protein interactions.Curr Opin Struct Biol. 2018 Jun;50:42-48. doi: 10.1016/j.sbi.2017.10.012. Epub 2017 Nov 5. Curr Opin Struct Biol. 2018. PMID: 29112911 Review.
-
Introducing the hypothome: a way to integrate predicted proteins in interactomes.Int J Bioinform Res Appl. 2014;10(6):647-52. doi: 10.1504/IJBRA.2014.065247. Int J Bioinform Res Appl. 2014. PMID: 25335568
-
Bacterial protein interaction networks: puzzle stones from solved complex structures add to a clearer picture.Integr Biol (Camb). 2011 Jun;3(6):645-52. doi: 10.1039/c0ib00023j. Epub 2011 May 17. Integr Biol (Camb). 2011. PMID: 21584322 Review.
Cited by
-
Prediction of Human-Plasmodium vivax Protein Associations From Heterogeneous Network Structures Based on Machine-Learning Approach.Bioinform Biol Insights. 2021 Jun 16;15:11779322211013350. doi: 10.1177/11779322211013350. eCollection 2021. Bioinform Biol Insights. 2021. PMID: 34188457 Free PMC article.
-
Computational Network Inference for Bacterial Interactomics.mSystems. 2022 Apr 26;7(2):e0145621. doi: 10.1128/msystems.01456-21. Epub 2022 Mar 30. mSystems. 2022. PMID: 35353009 Free PMC article. Review.
-
Proteome Data Improves Protein Function Prediction in the Interactome of Helicobacter pylori.Mol Cell Proteomics. 2018 May;17(5):961-973. doi: 10.1074/mcp.RA117.000474. Epub 2018 Feb 1. Mol Cell Proteomics. 2018. PMID: 29414760 Free PMC article.
-
Hot spot prediction in protein-protein interactions by an ensemble system.BMC Syst Biol. 2018 Dec 31;12(Suppl 9):132. doi: 10.1186/s12918-018-0665-8. BMC Syst Biol. 2018. PMID: 30598091 Free PMC article.
-
Combination of SAXS and Protein Painting Discloses the Three-Dimensional Organization of the Bacterial Cysteine Synthase Complex, a Potential Target for Enhancers of Antibiotic Action.Int J Mol Sci. 2019 Oct 21;20(20):5219. doi: 10.3390/ijms20205219. Int J Mol Sci. 2019. PMID: 31640223 Free PMC article.
References
-
- Abu-Farha M, Elisma F, Figeys D. Identification of protein-protein interactions by mass spectrometry coupled techniques. Adv Biochem Eng Biotechnol. 2008;110:67–80. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

