Immunoglobulin T from sea bass (Dicentrarchus labrax L.): molecular characterization, tissue localization and expression after nodavirus infection
- PMID: 28298204
- PMCID: PMC5353873
- DOI: 10.1186/s12867-017-0085-0
Immunoglobulin T from sea bass (Dicentrarchus labrax L.): molecular characterization, tissue localization and expression after nodavirus infection
Abstract
Background: Immunoglobulins (Igs) are fundamental components of the adaptive immune system of vertebrates, with the IgT/IgZ isotype specific of Teleosts. In this paper we describe the identification of an IgT heavy chain from the European sea bass (Dicentrarchus labrax L.), its molecular characterization and tissue mRNA localization by in situ hybridization.
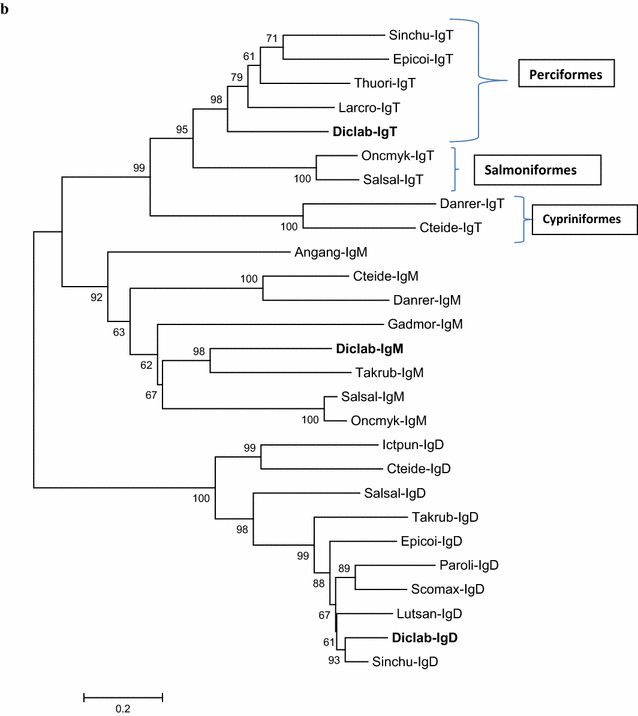
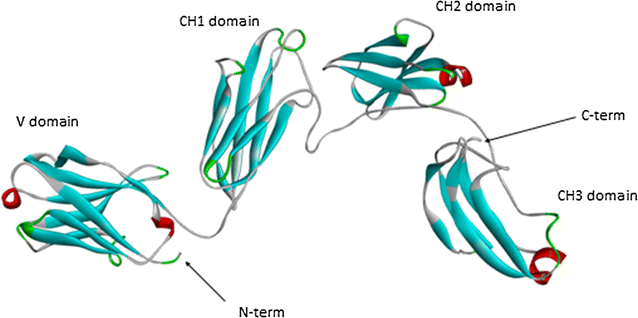
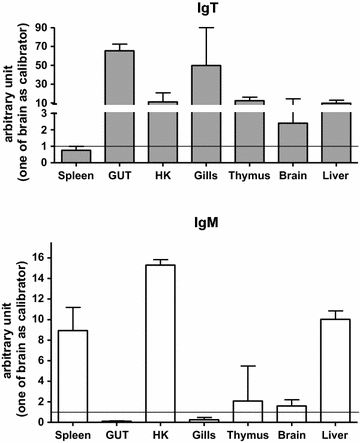
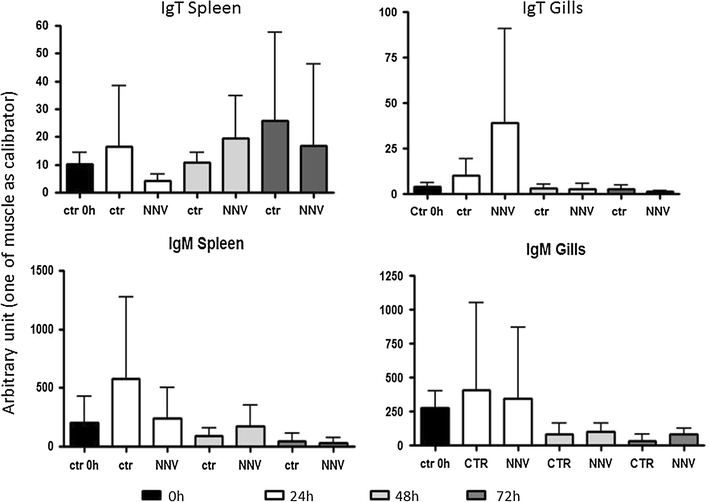
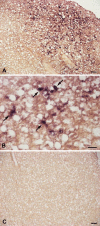
Results: Sea bass IgT consists of 552 aa (Accession Number KM410929) and it contains a putative 19 amino acids long signal peptide and one potential N-glycosylation site. The C-region consists of four CH domains; each contains the cysteine and tryptophan residues required for their correct folding. Based on the recent sequencing of sea bass genome, we have identified five different genomic contigs bearing exons unequivocally pertaining to IgT (CH2, CH3 and CH4), but none corresponded to a complete IgH locus as IgT sequences were found in the highly fragmented assembled genomic regions which could not be assigned to any major scaffold. The 3D structure of sea bass IgT has been modelled using the crystal structure of a mouse Ig gamma as a template, thus showing that the amino acid sequence is suitable for the expected topology referred to an immunoglobulin-like architecture. The basal expression of sea bass IgT and IgM in different organs has been analysed: gut and gills, important mucosal organs, showed high IgT transcripts levels and this was the first indication of the possible involvement of sea bass IgT in mucosal immune responses. Moreover, sea bass IgT expression increased in gills and spleen after infection with nodavirus, highlighting the importance of IgT in sea bass immune responses. In situ hybridization confirmed the presence of IgT transcripts in the gut and it revealed a differential expression along the intestinal tract, with a major expression in the posterior intestine, suggesting the hindgut as a site for the recruitment of IgT+ cells in this species. IgT transcripts were also found in gill filaments and parallel lamellae and, for the first time, we identified scattered IgT positive cells in the liver, with a strong signal in the hepatic parenchyma.
Conclusions: In conclusion, we performed a full molecular characterization of IgT in sea bass that points out its possible involvement in mucosal immune responses of this species.
Keywords: IgT; In situ hybridisation; Mucosal immunity; Sea bass; Tissue expression.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

