IRFinder: assessing the impact of intron retention on mammalian gene expression
- PMID: 28298237
- PMCID: PMC5353968
- DOI: 10.1186/s13059-017-1184-4
IRFinder: assessing the impact of intron retention on mammalian gene expression
Abstract
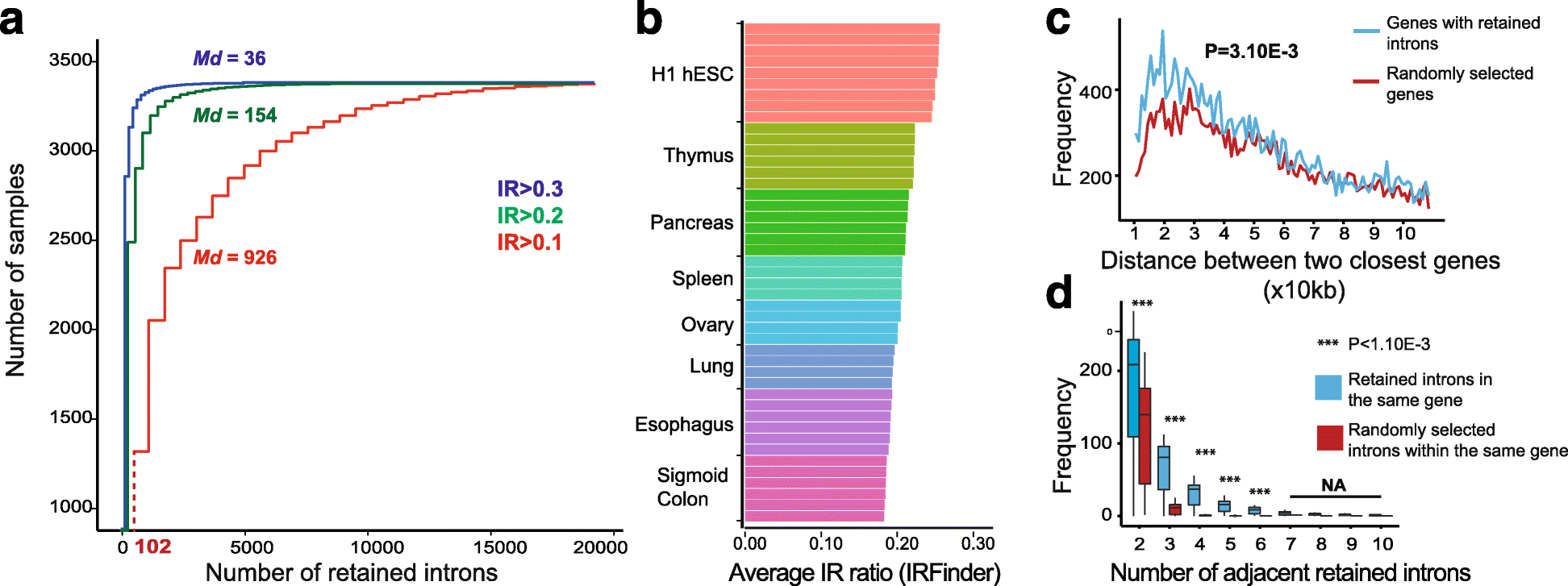
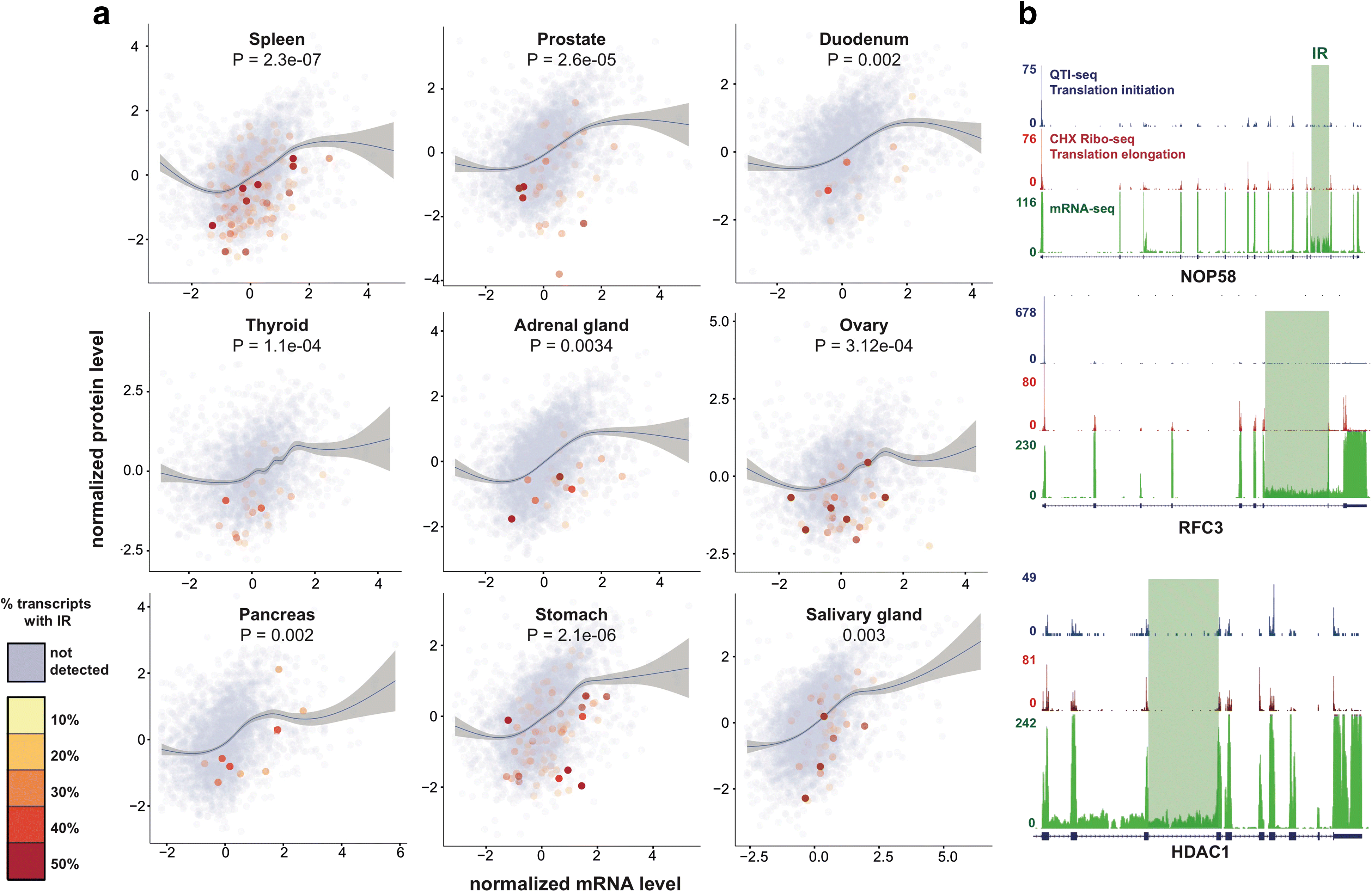
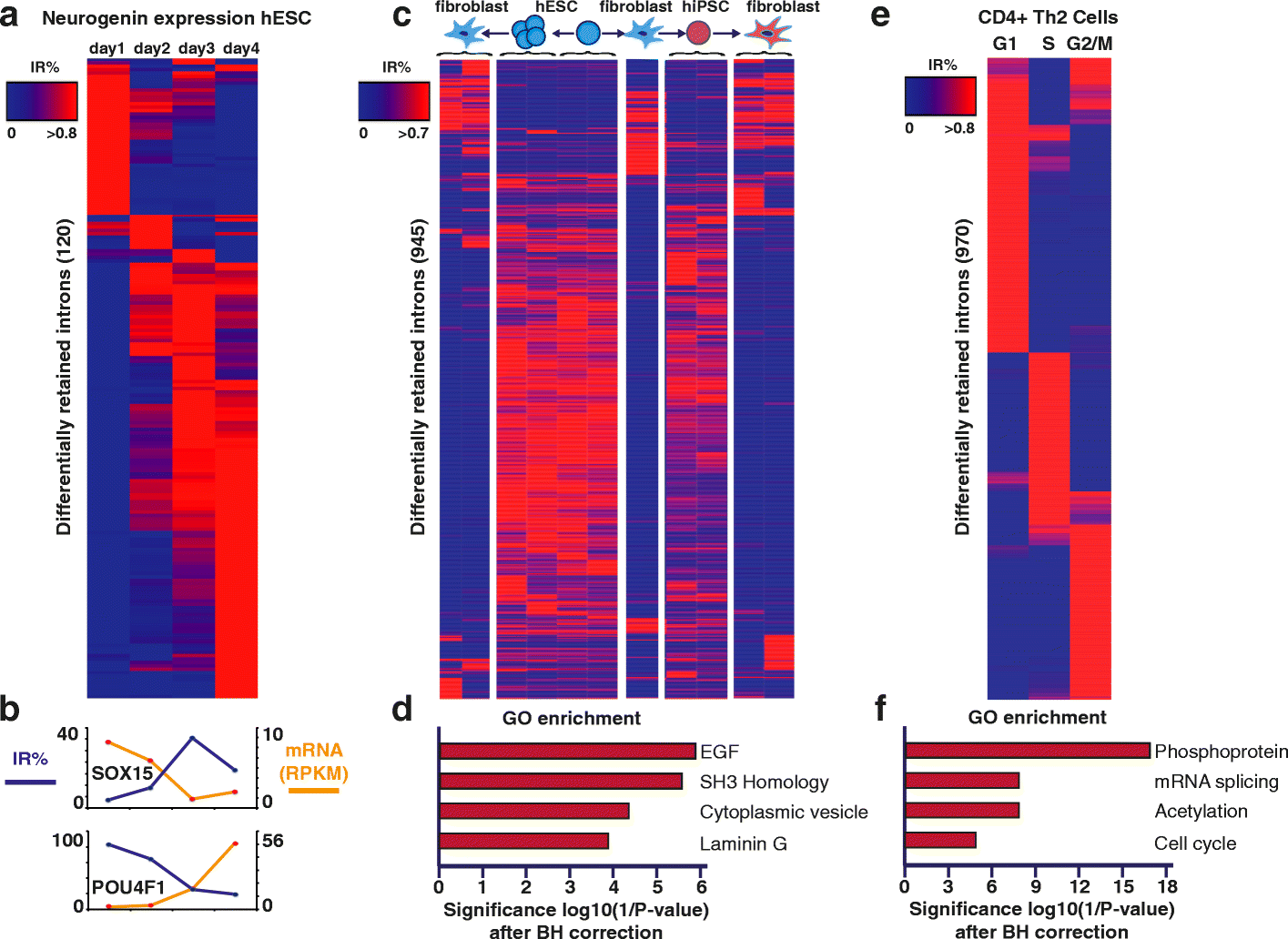
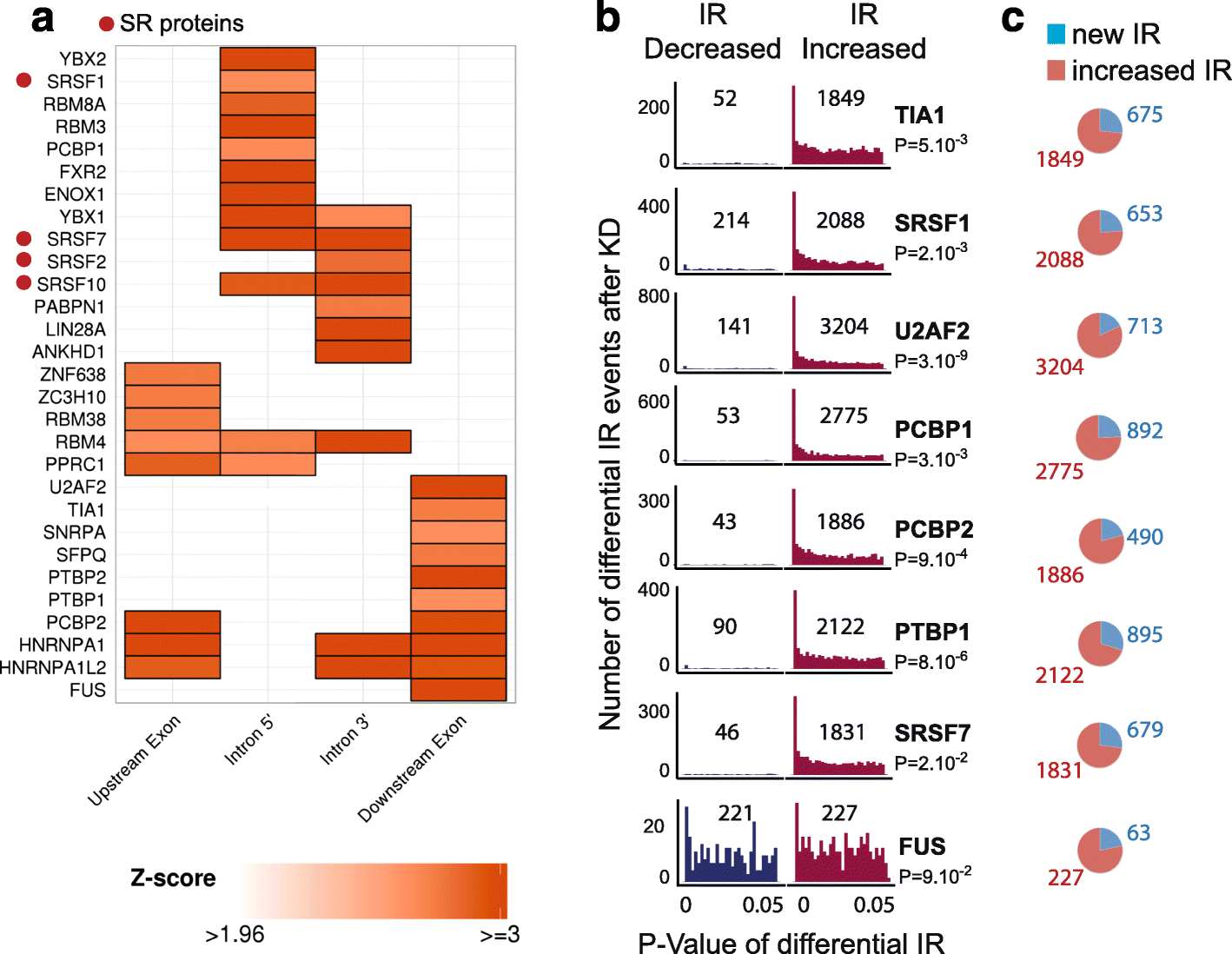
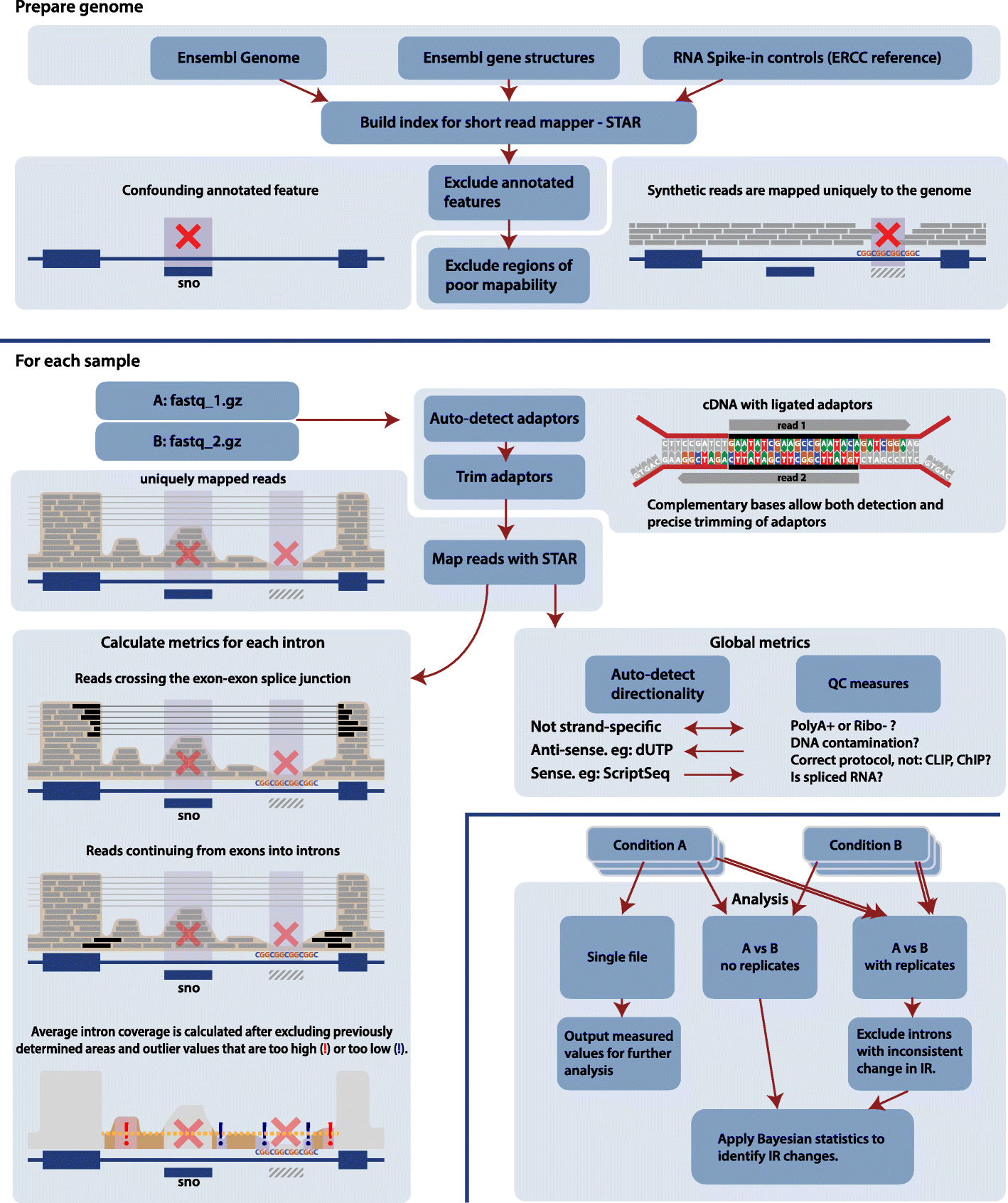
Intron retention (IR) occurs when an intron is transcribed into pre-mRNA and remains in the final mRNA. We have developed a program and database called IRFinder to accurately detect IR from mRNA sequencing data. Analysis of 2573 samples showed that IR occurs in all tissues analyzed, affects over 80% of all coding genes and is associated with cell differentiation and the cell cycle. Frequently retained introns are enriched for specific RNA binding protein sites and are often retained in clusters in the same gene. IR is associated with lower protein levels and intron-retaining transcripts that escape nonsense-mediated decay are not actively translated.
Keywords: Gene regulation; Intron retention; mRNA splicing.
Figures
Similar articles
-
IntEREst: intron-exon retention estimator.BMC Bioinformatics. 2018 Apr 11;19(1):130. doi: 10.1186/s12859-018-2122-5. BMC Bioinformatics. 2018. PMID: 29642843 Free PMC article.
-
The RNA-binding protein hnRNPLL induces a T cell alternative splicing program delineated by differential intron retention in polyadenylated RNA.Genome Biol. 2014 Jan 29;15(1):R26. doi: 10.1186/gb-2014-15-1-r26. Genome Biol. 2014. PMID: 24476532 Free PMC article.
-
A dynamic intron retention program enriched in RNA processing genes regulates gene expression during terminal erythropoiesis.Nucleic Acids Res. 2016 Jan 29;44(2):838-51. doi: 10.1093/nar/gkv1168. Epub 2015 Nov 3. Nucleic Acids Res. 2016. PMID: 26531823 Free PMC article.
-
Intron retention in mRNA: No longer nonsense: Known and putative roles of intron retention in normal and disease biology.Bioessays. 2016 Jan;38(1):41-9. doi: 10.1002/bies.201500117. Epub 2015 Nov 27. Bioessays. 2016. PMID: 26612485 Review.
-
The functional consequences of intron retention: alternative splicing coupled to NMD as a regulator of gene expression.Bioessays. 2014 Mar;36(3):236-43. doi: 10.1002/bies.201300156. Epub 2013 Dec 18. Bioessays. 2014. PMID: 24352796 Review.
Cited by
-
Sierra: discovery of differential transcript usage from polyA-captured single-cell RNA-seq data.Genome Biol. 2020 Jul 8;21(1):167. doi: 10.1186/s13059-020-02071-7. Genome Biol. 2020. PMID: 32641141 Free PMC article.
-
Direct Nanopore Sequencing of mRNA Reveals Landscape of Transcript Isoforms in Apicomplexan Parasites.mSystems. 2021 Mar 9;6(2):e01081-20. doi: 10.1128/mSystems.01081-20. mSystems. 2021. PMID: 33688018 Free PMC article.
-
Replicative aging in yeast involves dynamic intron retention patterns associated with mRNA processing/export and protein ubiquitination.Microb Cell. 2024 Feb 23;11:69-78. doi: 10.15698/mic2024.02.816. eCollection 2024. Microb Cell. 2024. PMID: 38414808 Free PMC article.
-
Identification of Alternative Polyadenylation in Cyanidioschyzon merolae Through Long-Read Sequencing of mRNA.Front Genet. 2022 Jan 28;12:818697. doi: 10.3389/fgene.2021.818697. eCollection 2021. Front Genet. 2022. PMID: 35154260 Free PMC article.
-
The variant rs77559646 associated with aggressive prostate cancer disrupts ANO7 mRNA splicing and protein expression.Hum Mol Genet. 2022 Jun 22;31(12):2063-2077. doi: 10.1093/hmg/ddac012. Hum Mol Genet. 2022. PMID: 35043958 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

