Staged induction of HIV-1 glycan-dependent broadly neutralizing antibodies
- PMID: 28298420
- PMCID: PMC5562350
- DOI: 10.1126/scitranslmed.aai7514
Staged induction of HIV-1 glycan-dependent broadly neutralizing antibodies
Abstract
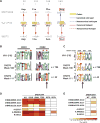
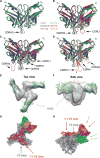
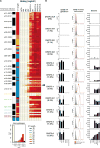
A preventive HIV-1 vaccine should induce HIV-1-specific broadly neutralizing antibodies (bnAbs). However, bnAbs generally require high levels of somatic hypermutation (SHM) to acquire breadth, and current vaccine strategies have not been successful in inducing bnAbs. Because bnAbs directed against a glycosylated site adjacent to the third variable loop (V3) of the HIV-1 envelope protein require limited SHM, the V3-glycan epitope is an attractive vaccine target. By studying the cooperation among multiple V3-glycan B cell lineages and their coevolution with autologous virus throughout 5 years of infection, we identify key events in the ontogeny of a V3-glycan bnAb. Two autologous neutralizing antibody lineages selected for virus escape mutations and consequently allowed initiation and affinity maturation of a V3-glycan bnAb lineage. The nucleotide substitution required to initiate the bnAb lineage occurred at a low-probability site for activation-induced cytidine deaminase activity. Cooperation of B cell lineages and an improbable mutation critical for bnAb activity defined the necessary events leading to breadth in this V3-glycan bnAb lineage. These findings may, in part, explain why initiation of V3-glycan bnAbs is rare, and suggest an immunization strategy for inducing similar V3-glycan bnAbs.
Copyright © 2017, American Association for the Advancement of Science.
Conflict of interest statement
Figures



References
-
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. - PMC - PubMed
-
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

