Speciation network in Laurasiatheria: retrophylogenomic signals
- PMID: 28298429
- PMCID: PMC5453332
- DOI: 10.1101/gr.210948.116
Speciation network in Laurasiatheria: retrophylogenomic signals
Abstract
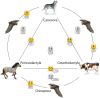
Rapid species radiation due to adaptive changes or occupation of new ecospaces challenges our understanding of ancestral speciation and the relationships of modern species. At the molecular level, rapid radiation with successive speciations over short time periods-too short to fix polymorphic alleles-is described as incomplete lineage sorting. Incomplete lineage sorting leads to random fixation of genetic markers and hence, random signals of relationships in phylogenetic reconstructions. The situation is further complicated when you consider that the genome is a mosaic of ancestral and modern incompletely sorted sequence blocks that leads to reconstructed affiliations to one or the other relative, depending on the fixation of their shared ancestral polymorphic alleles. The laurasiatherian relationships among Chiroptera, Perissodactyla, Cetartiodactyla, and Carnivora present a prime example for such enigmatic affiliations. We performed whole-genome screenings for phylogenetically diagnostic retrotransposon insertions involving the representatives bat (Chiroptera), horse (Perissodactyla), cow (Cetartiodactyla), and dog (Carnivora), and extracted among 162,000 preselected cases 102 virtually homoplasy-free, phylogenetically informative retroelements to draw a complete picture of the highly complex evolutionary relations within Laurasiatheria. All possible evolutionary scenarios received considerable retrotransposon support, leaving us with a network of affiliations. However, the Cetartiodactyla-Carnivora relationship as well as the basal position of Chiroptera and an ancestral laurasiatherian hybridization process did exhibit some very clear, distinct signals. The significant accordance of retrotransposon presence/absence patterns and flanking nucleotide changes suggest an important influence of mosaic genome structures in the reconstruction of species histories.
© 2017 Doronina et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- Avise JC, Robinson TJ. 2008. Hemiplasy: a new term in the lexicon of phylogenetics. Syst Biol 57: 503–507. - PubMed
-
- Bapteste E, van Iersel L, Janke A, Kelchner S, Kelk S, McInerney JO, Morrison DA, Nakhleh L, Steel M, Stougie L, et al. 2013. Networks: expanding evolutionary thinking. Trends Genet 29: 439–441. - PubMed
-
- Breen M, Bullerdiek J, Langford CF. 1999. The DAPI banded karyotype of the domestic dog (Canis familiaris) generated using chromosome-specific paint probes. Chromosome Res 7: 401–406. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
