Single-cell analysis reveals lineage segregation in early post-implantation mouse embryos
- PMID: 28298438
- PMCID: PMC5465504
- DOI: 10.1074/jbc.M117.780585
Single-cell analysis reveals lineage segregation in early post-implantation mouse embryos
Abstract
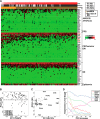
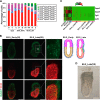
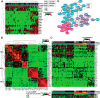
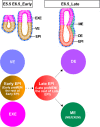
The mammalian post-implantation embryo has been extensively investigated at the tissue level. However, to unravel the molecular basis for the cell-fate plasticity and determination, it is essential to study the characteristics of individual cells. In particular, the individual definitive endoderm (DE) cells have not been characterized in vivo Here, we report gene expression patterns in single cells freshly isolated from mouse embryos on days 5.5 and 6.5. Initial transcriptome data from 124 single cells yielded signature genes for the epiblast, visceral endoderm, and extra-embryonic ectoderm and revealed a unique distribution pattern of fibroblast growth factor (FGF) ligands and receptors. Further analysis indicated that early-stage epiblast cells do not segregate into lineages of the major germ layers. Instead, some cells began to diverge from epiblast cells, displaying molecular features of the premesendoderm by expressing higher levels of mesendoderm markers and lower levels of Sox3 transcripts. Analysis of single-cell high-throughput quantitative RT-PCR data from 441 cells identified a late stage of the day 6.5 embryo in which mesoderm and DE cells emerge, with many of them coexpressing Oct4 and Gata6 Analysis of single-cell RNA-sequence data from 112 cells of the late-stage day 6.5 embryos revealed differentially expressed signaling genes and networks of transcription factors that might underlie the segregation of the mesoderm and DE lineages. Moreover, we discovered a subpopulation of mesoderm cells that possess molecular features of the extraembryonic mesoderm. This study provides fundamental insight into the molecular basis for lineage segregation in post-implantation mouse embryos.
Keywords: definitive endoderm; development; differentiation; embryo; extraembryonic mesoderm; gastrulation; gene regulation; mesoderm; single-cell RNA-Seq; transcriptomics.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Tam P. P., and Loebel D. A. (2007) Gene function in mouse embryogenesis: get set for gastrulation. Nat. Rev. Genet. 8, 368–381 - PubMed
-
- Guo G., Huss M., Tong G. Q., Wang C., Li Sun L., Clarke N. D., and Robson P. (2010) Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell 18, 675–685 - PubMed
-
- Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J., Huang J., Li M., Wu X., Wen L., Lao K., et al. (2013) Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1131–1139 - PubMed
-
- Ohnishi Y., Huber W., Tsumura A., Kang M., Xenopoulos P., Kurimoto K., Oleś A. K., Araúzo-Bravo M. J., Saitou M., Hadjantonakis A. K., and Hiiragi T. (2014) Cell-to-cell expression variability followed by signal reinforcement progressively segregates early mouse lineages. Nat. Cell Biol. 16, 27–37 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

