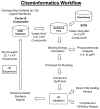
Identification of C3b-Binding Small-Molecule Complement Inhibitors Using Cheminformatics
- PMID: 28298523
- PMCID: PMC5417336
- DOI: 10.4049/jimmunol.1601932
Identification of C3b-Binding Small-Molecule Complement Inhibitors Using Cheminformatics
Abstract
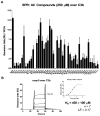
The complement system is an elegantly regulated biochemical cascade formed by the collective molecular recognition properties and proteolytic activities of more than two dozen membrane-bound or serum proteins. Complement plays diverse roles in human physiology, such as acting as a sentry against invading microorganisms, priming of the adaptive immune response, and removal of immune complexes. However, dysregulation of complement can serve as a trigger for a wide range of human diseases, which include autoimmune, inflammatory, and degenerative conditions. Despite several potential advantages of modulating complement with small-molecule inhibitors, small-molecule drugs are highly underrepresented in the current complement-directed therapeutics pipeline. In this study, we have employed a cheminformatics drug discovery approach based on the extensive structural and functional knowledge available for the central proteolytic fragment of the cascade, C3b. Using parallel in silico screening methodologies, we identified 45 small molecules that putatively bind C3b near ligand-guided functional hot spots. Surface plasmon resonance experiments resulted in the validation of seven dose-dependent C3b-binding compounds. Competition-based biochemical assays demonstrated the ability of several C3b-binding compounds to interfere with binding of the original C3b ligand that guided their discovery. In vitro assays of complement function identified a single complement inhibitory compound, termed cmp-5, and mechanistic studies of the cmp-5 inhibitory mode revealed it acts at the level of C5 activation. This study has led to the identification of a promising new class of C3b-binding small-molecule complement inhibitors and, to our knowledge, provides the first demonstration of cheminformatics-based, complement-directed drug discovery.
Copyright © 2017 by The American Association of Immunologists, Inc.
Figures


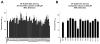
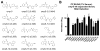


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

