Chemical approaches to targeted protein degradation through modulation of the ubiquitin-proteasome pathway
- PMID: 28298557
- PMCID: PMC5350610
- DOI: 10.1042/BCJ20160762
Chemical approaches to targeted protein degradation through modulation of the ubiquitin-proteasome pathway
Abstract
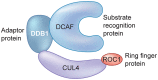
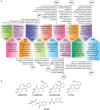
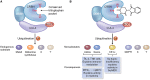
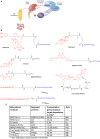
Manipulation of the ubiquitin-proteasome system to achieve targeted degradation of proteins within cells using chemical tools and drugs has the potential to transform pharmacological and therapeutic approaches in cancer and other diseases. An increased understanding of the molecular mechanism of thalidomide and its analogues following their clinical use has unlocked small-molecule modulation of the substrate specificity of the E3 ligase cereblon (CRBN), which in turn has resulted in the advancement of new immunomodulatory drugs (IMiDs) into the clinic. The degradation of multiple context-specific proteins by these pleiotropic small molecules provides a means to uncover new cell biology and to generate future drug molecules against currently undruggable targets. In parallel, the development of larger bifunctional molecules that bring together highly specific protein targets in complexes with CRBN, von Hippel-Lindau, or other E3 ligases to promote ubiquitin-dependent degradation has progressed to generate selective chemical compounds with potent effects in cells and in vivo models, providing valuable tools for biological target validation and with future potential for therapeutic use. In this review, we survey recent breakthroughs achieved in these two complementary methods and the discovery of new modes of direct and indirect engagement of target proteins with the proteasome. We discuss the experimental characterisation that validates the use of molecules that promote protein degradation as chemical tools, the preclinical and clinical examples disclosed to date, and the future prospects for this exciting area of chemical biology.
Keywords: chemical biology; chemical tools; ubiquitin ligases; ubiquitin–proteasome system.
© 2017 The Author(s).
Conflict of interest statement
Raj Chopra is a former employee of Celgene Corporation which has a commercial interest in IMiDs.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

