Proteolytic Cleavage of Bovine Adenovirus 3-Encoded pVIII
- PMID: 28298598
- PMCID: PMC5411589
- DOI: 10.1128/JVI.00211-17
Proteolytic Cleavage of Bovine Adenovirus 3-Encoded pVIII
Abstract
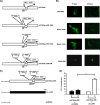
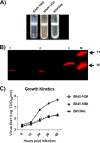
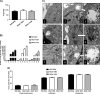
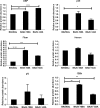
Proteolytic maturation involving cleavage of one nonstructural and six structural precursor proteins including pVIII by adenovirus protease is an important aspect of the adenovirus life cycle. The pVIII encoded by bovine adenovirus 3 (BAdV-3) is a protein of 216 amino acids and contains two potential protease cleavage sites. Here, we report that BAdV-3 pVIII is cleaved by adenovirus protease at both potential consensus protease cleavage sites. Usage of at least one cleavage site appears essential for the production of progeny BAdV-3 virions as glycine-to-alanine mutation of both protease cleavage sites appears lethal for the production of progeny virions. However, mutation of a single protease cleavage site of BAdV-3 pVIII significantly affects the efficient production of infectious progeny virions. Further analysis revealed no significant defect in endosome escape, genome replication, capsid formation, and virus assembly. Interestingly, cleavage of pVIII at both potential cleavage sites appears essential for the production of stable BAdV-3 virions as BAdV-3 expressing pVIII containing a glycine-to-alanine mutation of either of the potential cleavage sites is thermolabile, and this mutation leads to the production of noninfectious virions.IMPORTANCE Here, we demonstrated that the BAdV-3 adenovirus protease cleaves BAdV-3 pVIII at both potential protease cleavage sites. Although cleavage of pVIII at one of the two adenoviral protease cleavage sites is required for the production of progeny virions, the mutation of a single cleavage site of pVIII affects the efficient production of infectious progeny virions. Further analysis indicated that the mutation of a single protease cleavage site (glycine to alanine) of pVIII produces thermolabile virions, which leads to the production of noninfectious virions with disrupted capsids. We thus provide evidence about the requirement of proteolytic cleavage of pVIII for production of infectious progeny virions. We feel that our study has significantly advanced the understanding of the requirement of adenovirus protease cleavage of pVIII.
Keywords: BAdV-3; adenovirus protease; pVIII; proteolytic cleavage; thermolabile.
Copyright © 2017 American Society for Microbiology.
Figures







Similar articles
-
RETRACTED: Role of Protein VII in the Production of Infectious Bovine Adenovirus-3 Virion.Viruses. 2024 Aug 19;16(8):1323. doi: 10.3390/v16081323. Viruses. 2024. Retraction in: Viruses. 2025 Aug 19;17(8):1133. doi: 10.3390/v17081133. PMID: 39205297 Free PMC article. Retracted.
-
Conserved regions of bovine adenovirus-3 pVIII contain functional domains involved in nuclear localization and packaging in mature infectious virions.J Gen Virol. 2014 Aug;95(Pt 8):1743-1754. doi: 10.1099/vir.0.065763-0. Epub 2014 May 21. J Gen Virol. 2014. PMID: 24854002
-
A Single Maturation Cleavage Site in Adenovirus Impacts Cell Entry and Capsid Assembly.J Virol. 2015 Oct 21;90(1):521-32. doi: 10.1128/JVI.02014-15. Print 2016 Jan 1. J Virol. 2015. PMID: 26491163 Free PMC article.
-
Vaccinia virus proteolysis--a review.Rev Med Virol. 2006 May-Jun;16(3):187-202. doi: 10.1002/rmv.499. Rev Med Virol. 2006. PMID: 16710840 Free PMC article. Review.
-
Structure, function and dynamics in adenovirus maturation.Viruses. 2014 Nov 21;6(11):4536-70. doi: 10.3390/v6114536. Viruses. 2014. PMID: 25421887 Free PMC article. Review.
Cited by
-
Porcine Adenovirus Type 3 E3 Encodes a Structural Protein Essential for Capsid Stability and Production of Infectious Progeny Virions.J Virol. 2018 Sep 26;92(20):e00680-18. doi: 10.1128/JVI.00680-18. Print 2018 Oct 15. J Virol. 2018. PMID: 30068639 Free PMC article.
-
Regions of Bovine Adenovirus-3 Protein VII Involved in Interactions with Viral and Cellular Proteins.Viruses. 2024 May 5;16(5):732. doi: 10.3390/v16050732. Viruses. 2024. PMID: 38793614 Free PMC article.
-
RETRACTED: Role of Protein VII in the Production of Infectious Bovine Adenovirus-3 Virion.Viruses. 2024 Aug 19;16(8):1323. doi: 10.3390/v16081323. Viruses. 2024. Retraction in: Viruses. 2025 Aug 19;17(8):1133. doi: 10.3390/v17081133. PMID: 39205297 Free PMC article. Retracted.
-
Nuclear and Nucleolar Localization of Bovine Adenovirus-3 Protein V.Front Microbiol. 2021 Jan 6;11:579593. doi: 10.3389/fmicb.2020.579593. eCollection 2020. Front Microbiol. 2021. PMID: 33488533 Free PMC article.
-
Bovine Adenovirus-3 Tropism for Bovine Leukocyte Sub-Populations.Viruses. 2020 Dec 12;12(12):1431. doi: 10.3390/v12121431. Viruses. 2020. PMID: 33322850 Free PMC article.
References
-
- Berk AJ. 2007. Adenoviridae: the viruses and their replication, p 2355–2395. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

