RNA-Based Stable Isotope Probing Suggests Allobaculum spp. as Particularly Active Glucose Assimilators in a Complex Murine Microbiota Cultured In Vitro
- PMID: 28299315
- PMCID: PMC5337319
- DOI: 10.1155/2017/1829685
RNA-Based Stable Isotope Probing Suggests Allobaculum spp. as Particularly Active Glucose Assimilators in a Complex Murine Microbiota Cultured In Vitro
Abstract
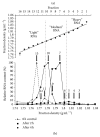
RNA-based stable isotope probing (RNA-SIP) and metabolic profiling were used to detect actively glucose-consuming bacteria in a complex microbial community obtained from a murine model system. A faeces-derived microbiota was incubated under anaerobic conditions for 0, 2, and 4 h with 40 mM [U13C]glucose. Isopycnic density gradient ultracentrifugation and fractionation of isolated RNA into labeled and unlabeled fractions followed by 16S rRNA sequencing showed a quick adaptation of the bacterial community in response to the added sugar, which was dominated by unclassified Lachnospiraceae species. Inspection of distinct fractions of isotope-labeled RNA revealed Allobaculum spp. as particularly active glucose utilizers in the system, as the corresponding RNA showed significantly higher proportions among the labeled RNA. With time, the labeled sugar was used by a wider spectrum of faecal bacteria. Metabolic profiling indicated rapid fermentation of [U13C]glucose, with lactate, acetate, and propionate being the principal 13C-labeled fermentation products, and suggested that "cross-feeding" occurred in the system. RNA-SIP combined with metabolic profiling of 13C-labeled products allowed insights into the microbial assimilation of a general model substrate, demonstrating the appropriateness of this technology to study assimilation processes of nutritionally more relevant substrates, for example, prebiotic carbohydrates, in the gut microbiota of mice as a model system.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

