Kilobase Pair Chromatin Fiber Contacts Promoted by Living-System-Like DNA Linker Length Distributions and Nucleosome Depletion
- PMID: 28299939
- PMCID: PMC6203935
- DOI: 10.1021/acs.jpcb.7b00998
Kilobase Pair Chromatin Fiber Contacts Promoted by Living-System-Like DNA Linker Length Distributions and Nucleosome Depletion
Abstract
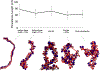
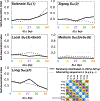
Nucleosome placement, or DNA linker length patterns, are believed to yield specific spatial features in chromatin fibers, but details are unknown. Here we examine by mesoscale modeling how kilobase (kb) range contacts and fiber looping depend on linker lengths ranging from 18 to 45 bp, with values modeled after living systems, including nucleosome free regions (NFRs) and gene encoding segments. We also compare artificial constructs with alternating versus randomly distributed linker lengths in the range of 18-72 bp. We show that nonuniform distributions with NFRs enhance flexibility and encourage kb-range contacts. NFRs between neighboring gene segments diminish short-range contacts between flanking nucleosomes, while enhancing kb-range contacts via hierarchical looping. We also demonstrate that variances in linker lengths enhance such contacts. In particular, moderate sized variations in fiber linker lengths (∼27 bp) encourage long-range contacts in randomly distributed linker length fibers. Our work underscores the importance of linker length patterns, alongside bound proteins, in biological regulation. Contacts formed by kb-range chromatin folding are crucial to gene activity. Because we find that special linker length distributions in living systems promote kb contacts, our work suggests ways to manipulate these patterns for regulation of gene activity.
Figures




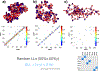
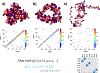
References
-
- Luger K; Mäder AW; Richmond RK; Sargent DF; Richmond TJ Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 1997, 389, 251–260. - PubMed
-
- Luger K; Hansen JC Nucleosome and chromatin fiber dynamics. Curr. Opin. Struct. Biol 2005, 15, 188–196. - PubMed
-
- Pombo A; Dillon N Three-dimensional genome architecture: players and mecha- nisms. Nature Rev. Mol. Cell Bio 2015, 16, 245–257. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

