The scaffold protein p140Cap limits ERBB2-mediated breast cancer progression interfering with Rac GTPase-controlled circuitries
- PMID: 28300085
- PMCID: PMC5357316
- DOI: 10.1038/ncomms14797
The scaffold protein p140Cap limits ERBB2-mediated breast cancer progression interfering with Rac GTPase-controlled circuitries
Erratum in
-
Author Correction: The scaffold protein p140Cap limits ERBB2-mediated breast cancer progression interfering with Rac GTPase-controlled circuitries.Nat Commun. 2018 Mar 30;9:16203. doi: 10.1038/ncomms16203. Nat Commun. 2018. PMID: 29600801 Free PMC article.
Abstract
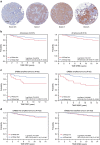
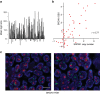
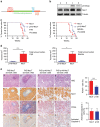
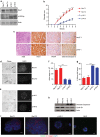
The docking protein p140Cap negatively regulates tumour cell features. Its relevance on breast cancer patient survival, as well as its ability to counteract relevant cancer signalling pathways, are not fully understood. Here we report that in patients with ERBB2-amplified breast cancer, a p140Cap-positive status associates with a significantly lower probability of developing a distant event, and a clear difference in survival. p140Cap dampens ERBB2-positive tumour cell progression, impairing tumour onset and growth in the NeuT mouse model, and counteracting epithelial mesenchymal transition, resulting in decreased metastasis formation. One major mechanism is the ability of p140Cap to interfere with ERBB2-dependent activation of Rac GTPase-controlled circuitries. Our findings point to a specific role of p140Cap in curbing the aggressiveness of ERBB2-amplified breast cancers and suggest that, due to its ability to impinge on specific molecular pathways, p140Cap may represent a predictive biomarker of response to targeted anti-ERBB2 therapies.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
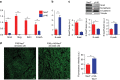

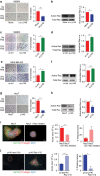
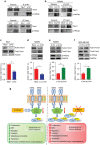
Similar articles
-
The p140Cap adaptor protein as a molecular hub to block cancer aggressiveness.Cell Mol Life Sci. 2021 Feb;78(4):1355-1367. doi: 10.1007/s00018-020-03666-w. Epub 2020 Oct 20. Cell Mol Life Sci. 2021. PMID: 33079227 Free PMC article. Review.
-
Dissecting the Shared and Context-Dependent Pathways Mediated by the p140Cap Adaptor Protein in Cancer and in Neurons.Front Cell Dev Biol. 2019 Oct 15;7:222. doi: 10.3389/fcell.2019.00222. eCollection 2019. Front Cell Dev Biol. 2019. PMID: 31681758 Free PMC article.
-
The SRCIN1/p140Cap adaptor protein negatively regulates the aggressiveness of neuroblastoma.Cell Death Differ. 2020 Feb;27(2):790-807. doi: 10.1038/s41418-019-0386-6. Epub 2019 Jul 8. Cell Death Differ. 2020. PMID: 31285546 Free PMC article.
-
p140Cap dual regulation of E-cadherin/EGFR cross-talk and Ras signalling in tumour cell scatter and proliferation.Oncogene. 2010 Jun 24;29(25):3677-90. doi: 10.1038/onc.2010.128. Epub 2010 May 10. Oncogene. 2010. PMID: 20453886
-
Tyrosine kinase signalling in breast cancer: tyrosine kinase-mediated signal transduction in transgenic mouse models of human breast cancer.Breast Cancer Res. 2000;2(3):211-6. doi: 10.1186/bcr56. Epub 2000 Apr 12. Breast Cancer Res. 2000. PMID: 11250712 Free PMC article. Review.
Cited by
-
The p140Cap adaptor protein as a molecular hub to block cancer aggressiveness.Cell Mol Life Sci. 2021 Feb;78(4):1355-1367. doi: 10.1007/s00018-020-03666-w. Epub 2020 Oct 20. Cell Mol Life Sci. 2021. PMID: 33079227 Free PMC article. Review.
-
Dissecting the Shared and Context-Dependent Pathways Mediated by the p140Cap Adaptor Protein in Cancer and in Neurons.Front Cell Dev Biol. 2019 Oct 15;7:222. doi: 10.3389/fcell.2019.00222. eCollection 2019. Front Cell Dev Biol. 2019. PMID: 31681758 Free PMC article.
-
Integrative Proteo-genomic Analysis to Construct CNA-protein Regulatory Map in Breast and Ovarian Tumors.Mol Cell Proteomics. 2019 Aug 9;18(8 suppl 1):S66-S81. doi: 10.1074/mcp.RA118.001229. Epub 2019 Jul 7. Mol Cell Proteomics. 2019. PMID: 31281117 Free PMC article.
-
Cooperative but distinct early co-signaling events originate from ERBB2 and ERBB1 receptors upon trastuzumab treatment in breast cancer cells.Oncotarget. 2017 May 8;8(36):60109-60122. doi: 10.18632/oncotarget.17686. eCollection 2017 Sep 1. Oncotarget. 2017. PMID: 28947957 Free PMC article.
-
p130Cas/BCAR1 and p140Cap/SRCIN1 Adaptors: The Yin Yang in Breast Cancer?Front Cell Dev Biol. 2021 Oct 11;9:729093. doi: 10.3389/fcell.2021.729093. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34708040 Free PMC article. Review.
References
-
- Siegel R. et al.. Cancer treatment and survivorship statistics, 2012. CA Cancer J. Clin. 62, 220–241 (2012). - PubMed
-
- Slamon D. J. et al.. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235, 177–182 (1987). - PubMed
-
- Hynes N. E. & MacDonald G. ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 21, 177–184 (2009). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

