Microbial Rhodopsins: Diversity, Mechanisms, and Optogenetic Applications
- PMID: 28301742
- PMCID: PMC5747503
- DOI: 10.1146/annurev-biochem-101910-144233
Microbial Rhodopsins: Diversity, Mechanisms, and Optogenetic Applications
Abstract
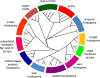
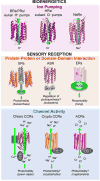
Microbial rhodopsins are a family of photoactive retinylidene proteins widespread throughout the microbial world. They are notable for their diversity of function, using variations of a shared seven-transmembrane helix design and similar photochemical reactions to carry out distinctly different light-driven energy and sensory transduction processes. Their study has contributed to our understanding of how evolution modifies protein scaffolds to create new protein chemistry, and their use as tools to control membrane potential with light is fundamental to optogenetics for research and clinical applications. We review the currently known functions and present more in-depth assessment of three functionally and structurally distinct types discovered over the past two years: (a) anion channelrhodopsins (ACRs) from cryptophyte algae, which enable efficient optogenetic neural suppression; (b) cryptophyte cation channelrhodopsins (CCRs), structurally distinct from the green algae CCRs used extensively for neural activation and from cryptophyte ACRs; and
Keywords: anion channelrhodopsins; cation channelrhodopsins; ion pumps; optogenetics; photosensors; retinylidene proteins.
(c) enzymerhodopsins, with light-gated guanylyl cyclase or kinase activity promising for optogenetic control of signal transduction.
Figures

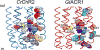



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

