Eukaryotic DNA Replication Fork
- PMID: 28301743
- PMCID: PMC5597965
- DOI: 10.1146/annurev-biochem-061516-044709
Eukaryotic DNA Replication Fork
Abstract
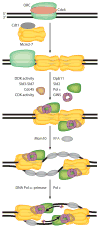
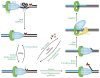
This review focuses on the biogenesis and composition of the eukaryotic DNA replication fork, with an emphasis on the enzymes that synthesize DNA and repair discontinuities on the lagging strand of the replication fork. Physical and genetic methodologies aimed at understanding these processes are discussed. The preponderance of evidence supports a model in which DNA polymerase ε (Pol ε) carries out the bulk of leading strand DNA synthesis at an undisturbed replication fork. DNA polymerases α and δ carry out the initiation of Okazaki fragment synthesis and its elongation and maturation, respectively. This review also discusses alternative proposals, including cellular processes during which alternative forks may be utilized, and new biochemical studies with purified proteins that are aimed at reconstituting leading and lagging strand DNA synthesis separately and as an integrated replication fork.
Keywords: CMG helicase; DNA polymerase; DNA primase; Okazaki fragment; replisome coordination.
Figures
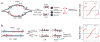


References
-
- Watson JD, Crick FH. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–67. - PubMed
-
- Okazaki R, Okazaki T, Sakabe K, Sugimoto K, Kainuma R, et al. In vivo mechanism of DNA chain growth. Cold Spring Harb Symp Quant Biol. 1968;33:129–43.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

