Implications of evolutionary engineering for growth and recombinant protein production in methanol-based growth media in the yeast Pichia pastoris
- PMID: 28302114
- PMCID: PMC5356285
- DOI: 10.1186/s12934-017-0661-5
Implications of evolutionary engineering for growth and recombinant protein production in methanol-based growth media in the yeast Pichia pastoris
Abstract
Background: Pichia pastoris is a widely used eukaryotic expression host for recombinant protein production. Adaptive laboratory evolution (ALE) has been applied in a wide range of studies in order to improve strains for biotechnological purposes. In this context, the impact of long-term carbon source adaptation in P. pastoris has not been addressed so far. Thus, we performed a pilot experiment in order to analyze the applicability and potential benefits of ALE towards improved growth and recombinant protein production in P. pastoris.
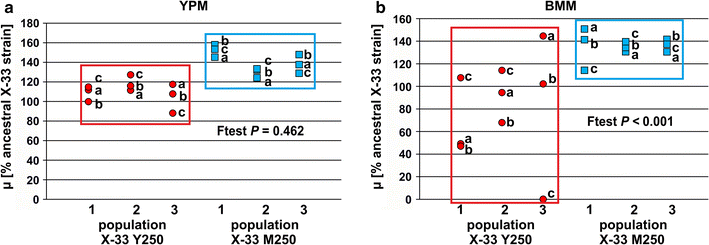
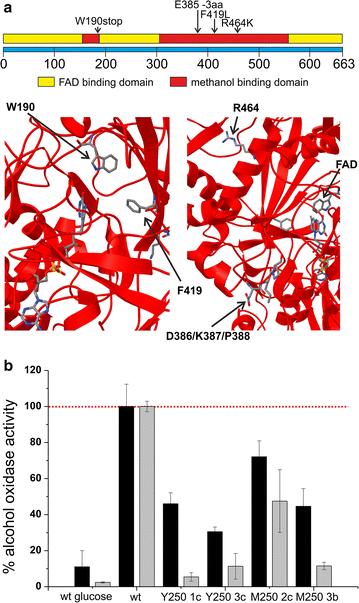
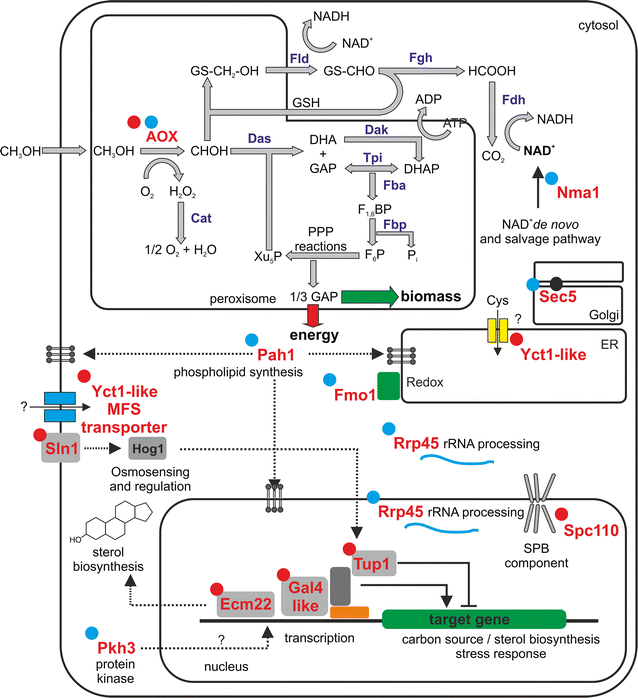
Results: Adaptation towards growth on methanol was performed in replicate cultures in rich and minimal growth medium for 250 generations. Increased growth rates on these growth media were observed at the population and single clone level. Evolved populations showed various degrees of growth advantages and trade-offs in non-evolutionary growth conditions. Genome resequencing revealed a wide variety of potential genetic targets associated with improved growth performance on methanol-based growth media. Alcohol oxidase represented a mutational hotspot since four out of seven evolved P. pastoris clones harbored mutations in this gene, resulting in decreased Aox activity, despite increased growth rates. Selected clones displayed strain-dependent variations for AOX-promoter based recombinant protein expression yield. One particularly interesting clone showed increased product titers ranging from a 2.5-fold increase in shake flask batch culture to a 1.8-fold increase during fed batch cultivation.
Conclusions: Our data indicate a complex correlation of carbon source, growth context and recombinant protein production. While similar experiments have already shown their potential in other biotechnological areas where microbes were evolutionary engineered for improved stress resistance and growth, the current dataset encourages the analysis of the potential of ALE for improved protein production in P. pastoris on a broader scale.
Keywords: Experimental evolution; Methanol; Pichia pastoris; Recombinant protein.
Figures
Similar articles
-
Functional recombinant protein is present in the pre-induction phases of Pichia pastoris cultures when grown in bioreactors, but not shake-flasks.Microb Cell Fact. 2014 Sep 4;13(1):127. doi: 10.1186/s12934-014-0127-y. Microb Cell Fact. 2014. PMID: 25186468 Free PMC article.
-
The influence of carbon sources on recombinant-human- growth-hormone production by Pichia pastoris is dependent on phenotype: a comparison of Muts and Mut+ strains.Biotechnol Appl Biochem. 2009 Mar;52(Pt 3):245-55. doi: 10.1042/BA20080057. Biotechnol Appl Biochem. 2009. PMID: 18754757
-
Biomarkers allow detection of nutrient limitations and respective supplementation for elimination in Pichia pastoris fed-batch cultures.Microb Cell Fact. 2017 Jul 11;16(1):117. doi: 10.1186/s12934-017-0730-9. Microb Cell Fact. 2017. PMID: 28693509 Free PMC article.
-
Carbon metabolism influenced for promoters and temperature used in the heterologous protein production using Pichia pastoris yeast.Braz J Microbiol. 2018 Nov;49 Suppl 1(Suppl 1):119-127. doi: 10.1016/j.bjm.2018.03.010. Epub 2018 May 19. Braz J Microbiol. 2018. PMID: 29858140 Free PMC article. Review.
-
Pichia pastoris Promoters.Methods Mol Biol. 2019;1923:97-112. doi: 10.1007/978-1-4939-9024-5_3. Methods Mol Biol. 2019. PMID: 30737736 Review.
Cited by
-
A one-step procedure for immobilising the thermostable carbonic anhydrase (SspCA) on the surface membrane of Escherichia coli.J Enzyme Inhib Med Chem. 2017 Dec;32(1):1120-1128. doi: 10.1080/14756366.2017.1355794. J Enzyme Inhib Med Chem. 2017. PMID: 28791907 Free PMC article.
-
Economical production of Pichia pastoris single cell protein from methanol at industrial pilot scale.Microb Cell Fact. 2023 Sep 28;22(1):198. doi: 10.1186/s12934-023-02198-9. Microb Cell Fact. 2023. PMID: 37770920 Free PMC article.
-
Improving Methanol Utilization by Reducing Alcohol Oxidase Activity and Adding Co-Substrate of Sodium Citrate in Pichia pastoris.J Fungi (Basel). 2023 Mar 29;9(4):422. doi: 10.3390/jof9040422. J Fungi (Basel). 2023. PMID: 37108877 Free PMC article.
-
Deciphering cell wall sensors enabling the construction of robust P. pastoris for single-cell protein production.Biotechnol Biofuels Bioprod. 2023 Nov 17;16(1):178. doi: 10.1186/s13068-023-02428-7. Biotechnol Biofuels Bioprod. 2023. PMID: 37978550 Free PMC article.
-
Methanol biotransformation toward high-level production of fatty acid derivatives by engineering the industrial yeast Pichia pastoris.Proc Natl Acad Sci U S A. 2022 Jul 19;119(29):e2201711119. doi: 10.1073/pnas.2201711119. Epub 2022 Jul 11. Proc Natl Acad Sci U S A. 2022. PMID: 35858340 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

