Genomic organization and recombination analysis of a porcine sapovirus identified from a piglet with diarrhea in China
- PMID: 28302145
- PMCID: PMC5356244
- DOI: 10.1186/s12985-017-0729-1
Genomic organization and recombination analysis of a porcine sapovirus identified from a piglet with diarrhea in China
Abstract
Background: Sapovirus (SaV), a member of the family Caliciviridae, is an etiologic agent of gastroenteritis in humans and pigs. To date, both intra- and inter-genogroup recombinant strains have been reported in many countries except for China. Here, we report an intra-genogroup recombination of porcine SaV identified from a piglet with diarrhea of China.
Methods: A fecal sample from a 15-day-old piglet with diarrhea was collected from Shanghai, China. Common agents of gastroenteritis including porcine circovirus type 2, porcine rotavirus, porcine transmissible gastroenteritis virus, porcine SaV, porcine norovirus, and porcine epidemic diarrhea virus were detected by RT-PCR or PCR method. The complete genome of porcine SaV was then determined by RT-PCR method. Phylogenetic analyses based on the structural region and nonstructural (NS) region were carried out to group this SaV strain, and it was divided into different genotypes based on these two regions. Recombination analysis based on the genomic sequence was further performed to confirm this recombinant event and locate the breakpoint.
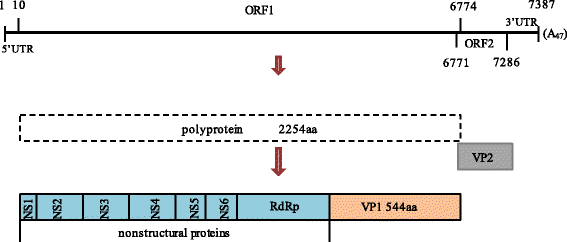
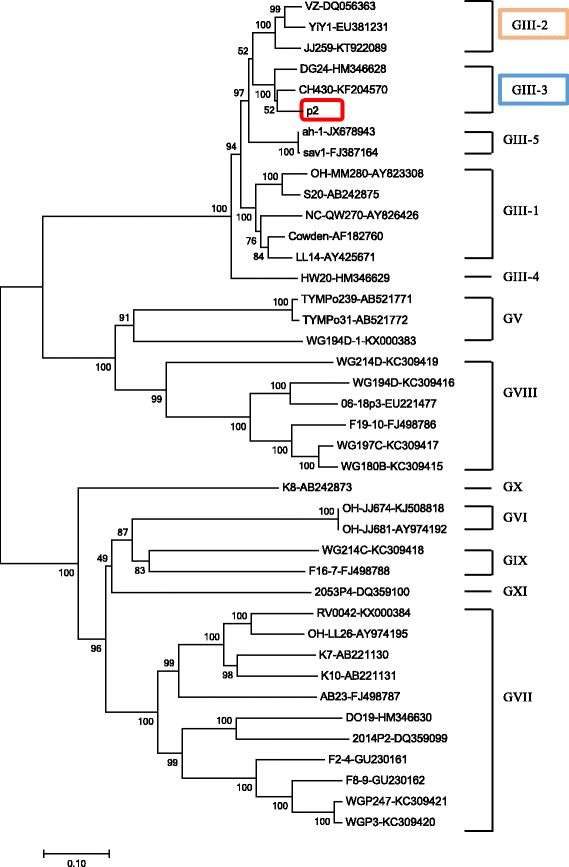
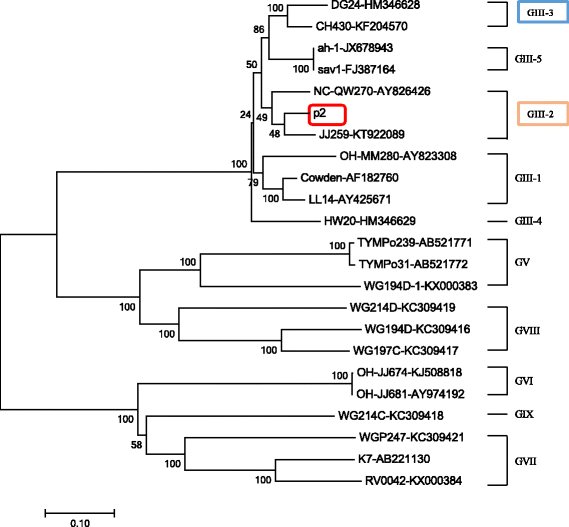
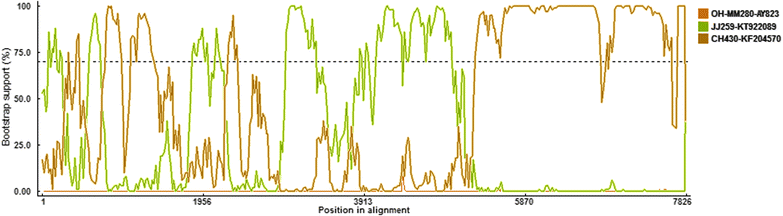
Results: All of the agents showed negative results except for SaV. Analysis of the complete genome sequence showed that this strain was 7387 nt long with two ORFs and belonged to SaV GIII. Phylogenetic analyses of the structural region (complete VP1 nucleotide sequences) grouped this strain into GIII-3, whereas of the nonstructural region (RdRp nucleotide sequences) grouped this strain into GIII-2. Recombination analysis based on the genomic sequence confirmed this recombinant event and identified two parental strains that were JJ259 (KT922089, GIII-2) and CH430 (KF204570, GIII-3). The breakpoint located at position 5139 nt of the genome (RdRp-capsid junction region). Etiologic analysis showed the fecal sample was negative with the common agents of gastroenteritis, except for porcine SaV, which suggested that this recombinant strain might lead to this piglet diarrhea.
Conclusions: P2 strain was an intra-genogroup recombinant porcine SaV. To the best of our knowledge, this study would be the first report that intra-genogroup recombination of porcine SaV infection was identified in pig herd in China.
Keywords: Genome organization; Porcine sapovirus; Recombination.
Figures
Similar articles
-
High genetic diversity in RdRp gene of Brazilian porcine sapovirus strains.Vet Microbiol. 2008 Sep 18;131(1-2):185-91. doi: 10.1016/j.vetmic.2008.02.021. Epub 2008 Mar 4. Vet Microbiol. 2008. PMID: 18403136
-
Identification of genetic diversity of porcine Norovirus and Sapovirus in Korea.Virus Genes. 2011 Jun;42(3):394-401. doi: 10.1007/s11262-011-0588-6. Epub 2011 Mar 3. Virus Genes. 2011. PMID: 21369826
-
Genetic diversity and recombination of porcine sapoviruses.J Clin Microbiol. 2005 Dec;43(12):5963-72. doi: 10.1128/JCM.43.12.5963-5972.2005. J Clin Microbiol. 2005. PMID: 16333083 Free PMC article.
-
Porcine sapoviruses: Pathogenesis, epidemiology, genetic diversity, and diagnosis.Virus Res. 2020 Sep;286:198025. doi: 10.1016/j.virusres.2020.198025. Epub 2020 May 26. Virus Res. 2020. PMID: 32470356 Free PMC article. Review.
-
Human sapoviruses: genetic diversity, recombination, and classification.Rev Med Virol. 2007 Mar-Apr;17(2):133-41. doi: 10.1002/rmv.533. Rev Med Virol. 2007. PMID: 17340567 Review.
Cited by
-
High Genetic Diversity of Porcine Sapovirus From Diarrheic Piglets in Yunnan Province, China.Front Vet Sci. 2022 Jul 7;9:854905. doi: 10.3389/fvets.2022.854905. eCollection 2022. Front Vet Sci. 2022. PMID: 35873674 Free PMC article.
-
Metagenomic Analysis of RNA Fraction Reveals the Diversity of Swine Oral Virome on South African Backyard Swine Farms in the uMgungundlovu District of KwaZulu-Natal Province.Pathogens. 2022 Aug 17;11(8):927. doi: 10.3390/pathogens11080927. Pathogens. 2022. PMID: 36015047 Free PMC article.
-
A novel recombinant porcine sapovirus infection in piglets with diarrhea in Shandong Province, China, 2022.Braz J Microbiol. 2023 Jun;54(2):1309-1314. doi: 10.1007/s42770-023-00963-x. Epub 2023 Apr 10. Braz J Microbiol. 2023. PMID: 37036658 Free PMC article.
-
Development of a Multiplex RT-PCR Assay for Simultaneous Detection of Four Potential Zoonotic Swine RNA Viruses.Vet Sci. 2022 Apr 7;9(4):176. doi: 10.3390/vetsci9040176. Vet Sci. 2022. PMID: 35448674 Free PMC article.
-
Genomic Evolution and Selective Pressure Analysis of a Novel Porcine Sapovirus in Shanghai, China.Microorganisms. 2024 Mar 12;12(3):569. doi: 10.3390/microorganisms12030569. Microorganisms. 2024. PMID: 38543620 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

