Identification of residues important for the activity of aldehyde-deformylating oxygenase through investigation into the structure-activity relationship
- PMID: 28302170
- PMCID: PMC5356278
- DOI: 10.1186/s12896-017-0351-8
Identification of residues important for the activity of aldehyde-deformylating oxygenase through investigation into the structure-activity relationship
Abstract
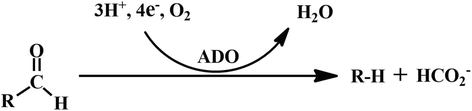
Background: Aldehyde-deformylating oxygenase (ADO) is a key enzyme involved in the biosynthetic pathway of fatty alk(a/e)nes in cyanobacteria. However, cADO (cyanobacterial ADO) showed extreme low activity with the k cat value below 1 min-1, which would limit its application in biofuel production. To identify the activity related key residues of cADO is urgently required.
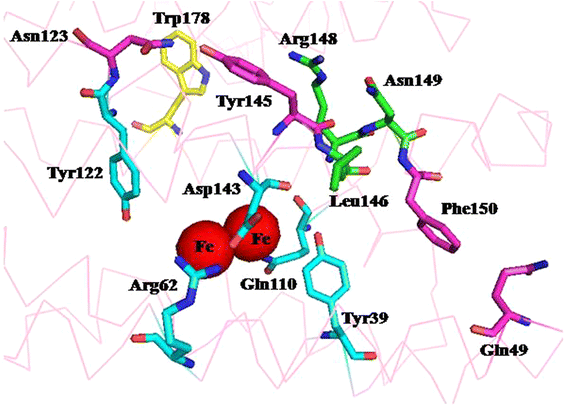
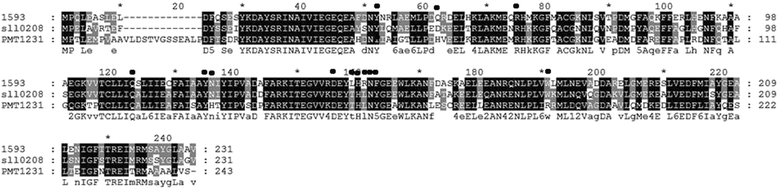
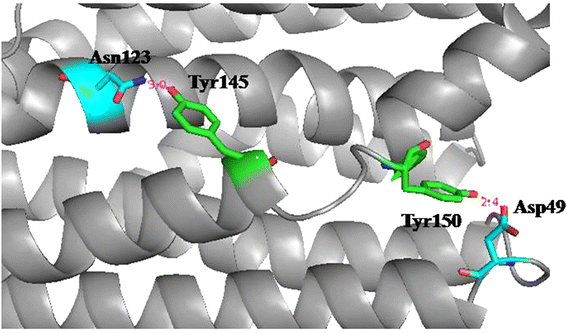
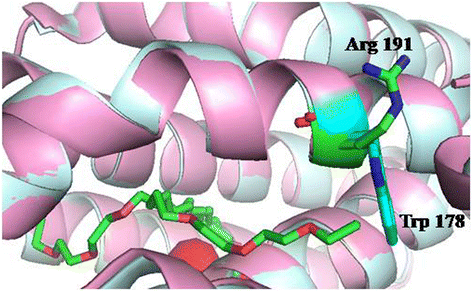
Results: The amino acid residues which might affect cADO activity were identified based on the crystal structures and sequence alignment of cADOs, including the residues close to the di-iron center (Tyr39, Arg62, Gln110, Tyr122, Asp143 of cADO-1593), the protein surface (Trp 178 of cADO-1593), and those involved in two important hydrogen bonds (Gln49, Asn123 of cADO-1593, and Asp49, Asn123 of cADO-sll0208) and in the oligopeptide whose conformation changed in the absence of the di-iron center (Leu146, Asn149, Phe150 of cADO-1593, and Thr146, Leu148, Tyr150 of cADO-sll0208). The variants of cADO-1593 from Synechococcus elongatus PCC7942 and cADO-sll0208 from Synechocystis sp. PCC6803 were constructed, overexpressed, purified and kinetically characterized. The k cat values of L146T, Q49H/N123H/F150Y and W178R of cADO-1593 and L148R of cADO-sll0208 were increased by more than two-fold, whereas that of R62A dropped by 91.1%. N123H, Y39F and D143A of cADO-1593, and Y150F of cADO-sll0208 reduced activities by ≤ 20%.
Conclusions: Some important amino acids, which exerted some effects on cADO activity, were identified. Several enzyme variants exhibited greatly reduced activity, while the k cat values of several mutants are more than two-fold higher than the wild type. This study presents the report on the relationship between amino acid residues and enzyme activity of cADOs, and the information will provide a guide for enhancement of cADO activity through protein engineering.
Keywords: Aldehyde-deformylating oxygenase; Fatty alk(a/e)ne; Site-directed mutagenesis; Structure-activity relationship; Synechococcus elongatus PCC7942; Synechocystis sp. PCC6803.
Figures
Similar articles
-
Structure-oriented substrate specificity engineering of aldehyde-deformylating oxygenase towards aldehydes carbon chain length.Biotechnol Biofuels. 2016 Aug 31;9(1):185. doi: 10.1186/s13068-016-0596-9. eCollection 2016. Biotechnol Biofuels. 2016. PMID: 27588038 Free PMC article.
-
Structural insights into the catalytic mechanism of aldehyde-deformylating oxygenases.Protein Cell. 2015 Jan;6(1):55-67. doi: 10.1007/s13238-014-0108-2. Epub 2014 Dec 9. Protein Cell. 2015. PMID: 25482408 Free PMC article.
-
Crystal structures of aldehyde deformylating oxygenase from Limnothrix sp. KNUA012 and Oscillatoria sp. KNUA011.Biochem Biophys Res Commun. 2016 Aug 26;477(3):395-400. doi: 10.1016/j.bbrc.2016.06.090. Epub 2016 Jun 18. Biochem Biophys Res Commun. 2016. PMID: 27329814
-
Cyanobacterial aldehyde deformylating oxygenase: Structure, function, and potential in biofuels production.Int J Biol Macromol. 2020 Dec 1;164:3155-3162. doi: 10.1016/j.ijbiomac.2020.08.162. Epub 2020 Aug 22. Int J Biol Macromol. 2020. PMID: 32841666 Review.
-
Cyanobacterial Enzymes for Bioalkane Production.Adv Exp Med Biol. 2018;1080:119-154. doi: 10.1007/978-981-13-0854-3_6. Adv Exp Med Biol. 2018. PMID: 30091094 Review.
Cited by
-
Ferritin-Like Proteins: A Conserved Core for a Myriad of Enzyme Complexes.Subcell Biochem. 2022;99:109-153. doi: 10.1007/978-3-031-00793-4_4. Subcell Biochem. 2022. PMID: 36151375
-
Recent advances in the improvement of cyanobacterial enzymes for bioalkane production.Microb Cell Fact. 2022 Dec 12;21(1):256. doi: 10.1186/s12934-022-01981-4. Microb Cell Fact. 2022. PMID: 36503511 Free PMC article. Review.
-
A consensus-guided approach yields a heat-stable alkane-producing enzyme and identifies residues promoting thermostability.J Biol Chem. 2018 Jun 15;293(24):9148-9161. doi: 10.1074/jbc.RA117.000639. Epub 2018 Apr 9. J Biol Chem. 2018. PMID: 29632075 Free PMC article.
-
Identification of non-conserved residues essential for improving the hydrocarbon-producing activity of cyanobacterial aldehyde-deformylating oxygenase.Biotechnol Biofuels. 2019 Apr 17;12:89. doi: 10.1186/s13068-019-1409-8. eCollection 2019. Biotechnol Biofuels. 2019. PMID: 31015863 Free PMC article.
-
Directed evolution of hydrocarbon-producing enzymes.Biotechnol Biofuels Bioprod. 2025 Aug 12;18(1):91. doi: 10.1186/s13068-025-02689-4. Biotechnol Biofuels Bioprod. 2025. PMID: 40796908 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

