Causal role for inheritance of H3K27me3 in maintaining the OFF state of a Drosophila HOX gene
- PMID: 28302795
- PMCID: PMC5595140
- DOI: 10.1126/science.aai8236
Causal role for inheritance of H3K27me3 in maintaining the OFF state of a Drosophila HOX gene
Abstract
Many eukaryotic cells can respond to transient environmental or developmental stimuli with heritable changes in gene expression that are associated with nucleosome modifications. However, it remains uncertain whether modified nucleosomes play a causal role in transmitting such epigenetic memories, as opposed to controlling or merely reflecting transcriptional states inherited by other means. Here, we provide in vivo evidence that H3K27 trimethylated nucleosomes, once established at a repressed Drosophila HOX gene, remain heritably associated with that gene and can carry the memory of the silenced state through multiple rounds of replication, even when the capacity to copy the H3K27me3 mark to newly incorporated nucleosomes is diminished or abolished. Hence, in this context, the inheritance of H3K27 trimethylation conveys epigenetic memory.
Copyright © 2017, American Association for the Advancement of Science.
Figures

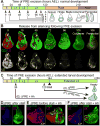
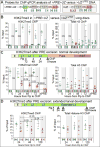
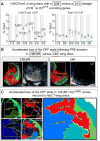

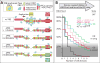
Comment in
-
Passing epigenetic silence to the next generation.Science. 2017 Apr 7;356(6333):28-29. doi: 10.1126/science.aan1493. Science. 2017. PMID: 28385971 No abstract available.
References
-
- Ptashne M. Principles of a switch. Nat Chem Biol. 2011;7:484–487. - PubMed
-
- Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits. Chapman & Hall/CRC Press/Taylor & Fracis; Boca Raton, FL: 2006.
-
- Steffen PA, Ringrose L. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nature Reviews Molecular Cell Biology. 2014;15:340–356. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

