Ligand co-crystallization of aminoacyl-tRNA synthetases from infectious disease organisms
- PMID: 28303005
- PMCID: PMC5428304
- DOI: 10.1038/s41598-017-00367-6
Ligand co-crystallization of aminoacyl-tRNA synthetases from infectious disease organisms
Abstract
Aminoacyl-tRNA synthetases (aaRSs) charge tRNAs with their cognate amino acid, an essential precursor step to loading of charged tRNAs onto the ribosome and addition of the amino acid to the growing polypeptide chain during protein synthesis. Because of this important biological function, aminoacyl-tRNA synthetases have been the focus of anti-infective drug development efforts and two aaRS inhibitors have been approved as drugs. Several researchers in the scientific community requested aminoacyl-tRNA synthetases to be targeted in the Seattle Structural Genomics Center for Infectious Disease (SSGCID) structure determination pipeline. Here we investigate thirty-one aminoacyl-tRNA synthetases from infectious disease organisms by co-crystallization in the presence of their cognate amino acid, ATP, and/or inhibitors. Crystal structures were determined for a CysRS from Borrelia burgdorferi bound to AMP, GluRS from Borrelia burgdorferi and Burkholderia thailandensis bound to glutamic acid, a TrpRS from the eukaryotic pathogen Encephalitozoon cuniculi bound to tryptophan, a HisRS from Burkholderia thailandensis bound to histidine, and a LysRS from Burkholderia thailandensis bound to lysine. Thus, the presence of ligands may promote aaRS crystallization and structure determination. Comparison with homologous structures shows conformational flexibility that appears to be a recurring theme with this enzyme class.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
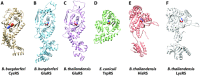
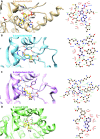
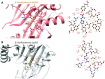
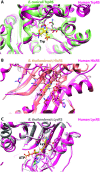
Similar articles
-
Recombinant expression and purification of wheat aminoacyl-tRNA synthetases and IRES-dependent polypeptide synthesis with homogeneously derived purified factors.Biochimie. 2025 Sep;236:30-44. doi: 10.1016/j.biochi.2025.06.010. Epub 2025 Jun 21. Biochimie. 2025. PMID: 40550411
-
Human disease-causing missense genetic variants are enriched in the evolutionarily ancient domains of the cytosolic aminoacyl-tRNA synthetase proteins.IUBMB Life. 2025 Jan;77(1):e2932. doi: 10.1002/iub.2932. IUBMB Life. 2025. PMID: 39710895 Free PMC article.
-
Structural Enzymology, Phylogenetics, Differentiation, and Symbolic Reflexivity at the Dawn of Biology.Genome Biol Evol. 2025 May 30;17(6):evaf095. doi: 10.1093/gbe/evaf095. Genome Biol Evol. 2025. PMID: 40476352 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Direct and quantitative analysis of tRNA acylation using intact tRNA liquid chromatography-mass spectrometry.Nat Protoc. 2025 May;20(5):1246-1274. doi: 10.1038/s41596-024-01086-9. Epub 2025 Jan 6. Nat Protoc. 2025. PMID: 39762443 Review.
Cited by
-
Crystal structure of glutamyl-tRNA synthetase from Helicobacter pylori.Acta Crystallogr F Struct Biol Commun. 2024 Dec 1;80(Pt 12):335-40. doi: 10.1107/S2053230X24011099. Online ahead of print. Acta Crystallogr F Struct Biol Commun. 2024. PMID: 39601417 Free PMC article.
-
The mechanism of lineage-specific tRNA recognition by bacterial tryptophanyl-tRNA synthetase and its implications for inhibitor discovery.Nucleic Acids Res. 2025 May 22;53(10):gkaf466. doi: 10.1093/nar/gkaf466. Nucleic Acids Res. 2025. PMID: 40464690 Free PMC article.
-
Crystal structures of glutamyl-tRNA synthetase from Elizabethkingia anopheles and E. meningosepticum.Acta Crystallogr F Struct Biol Commun. 2022 Aug 1;78(Pt 8):306-312. doi: 10.1107/S2053230X22007555. Epub 2022 Jul 28. Acta Crystallogr F Struct Biol Commun. 2022. PMID: 35924598 Free PMC article.
-
Lysyl-tRNA Synthetase from Pseudomonas aeruginosa: Characterization and Identification of Inhibitory Compounds.SLAS Discov. 2020 Jan;25(1):57-69. doi: 10.1177/2472555219873095. Epub 2019 Sep 9. SLAS Discov. 2020. PMID: 31498734 Free PMC article.
-
3-Dimensional architecture of the human multi-tRNA synthetase complex.Nucleic Acids Res. 2020 Sep 4;48(15):8740-8754. doi: 10.1093/nar/gkaa569. Nucleic Acids Res. 2020. PMID: 32644155 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

