Development of SyneBrick Vectors As a Synthetic Biology Platform for Gene Expression in Synechococcus elongatus PCC 7942
- PMID: 28303150
- PMCID: PMC5332412
- DOI: 10.3389/fpls.2017.00293
Development of SyneBrick Vectors As a Synthetic Biology Platform for Gene Expression in Synechococcus elongatus PCC 7942
Abstract
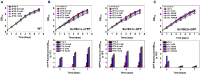
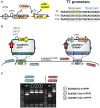
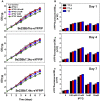
Cyanobacteria are oxygenic photosynthetic prokaryotes that are able to assimilate CO2 using solar energy and water. Metabolic engineering of cyanobacteria has suggested the possibility of direct CO2 conversion to value-added chemicals. However, engineering of cyanobacteria has been limited due to the lack of various genetic tools for expression and control of multiple genes to reconstruct metabolic pathways for biochemicals from CO2. Thus, we developed SyneBrick vectors as a synthetic biology platform for gene expression in Synechococcus elongatus PCC 7942 as a model cyanobacterium. The SyneBrick chromosomal integration vectors provide three inducible expression systems to control gene expression and three neutral sites for chromosomal integrations. Using a SyneBrick vector, LacI-regulated gene expression led to 24-fold induction of the eYFP reporter gene with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) inducer in S. elongatus PCC 7942 under 5% (v/v) CO2. TetR-regulated gene expression led to 19-fold induction of the GFP gene when 100 nM anhydrotetracycline (aTc) inducer was used. Gene expression decreased after 48 h due to degradation of aTc under light. T7 RNA polymerase-based gene expression resulted in efficient expression with a lower IPTG concentration than a previously developed pTrc promoter. A library of T7 promoters can be used for tunable gene expression. In summary, SyneBrick vectors were developed as a synthetic biology platform for gene expression in S. elongatus PCC 7942. These results will accelerate metabolic engineering of biosolar cell factories through expressing and controlling multiple genes of interest.
Keywords: SyneBrick vectors; Synechococcus elongatus PCC 7942; T7 promoters; gene expression; synthetic biology.
Figures

References
-
- Chwa J. W., Kim W. J., Sim S. J., Um Y., Woo H. M. (2016). Engineering of a modular and synthetic phosphoketolase pathway for photosynthetic production of acetone from CO2 in Synechococcus elongatus PCC 7942 under light and aerobic condition. Plant Biotechnol. J. 14, 1768–1776. 10.1111/pbi.12536 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

