Agreement between an in-house replication competent and a reference replication defective recombinant virus assay for measuring phenotypic resistance to HIV-1 protease, reverse transcriptase, and integrase inhibitors
- PMID: 28303602
- PMCID: PMC6816933
- DOI: 10.1002/jcla.22206
Agreement between an in-house replication competent and a reference replication defective recombinant virus assay for measuring phenotypic resistance to HIV-1 protease, reverse transcriptase, and integrase inhibitors
Abstract
Background: Although clinical management of drug resistance is routinely based on genotypic methods, phenotypic assays remain necessary for the characterization of novel HIV-1 inhibitors, particularly against common drug-resistant variants. We describe the development and assessment of the performance of a recombinant virus assay for measuring HIV-1 susceptibility to protease (PR), reverse transcriptase (RT), and integrase (IN) inhibitors.
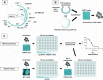
Methods: The system is based on the creation of replication-competent chimeric viruses through homologous recombination between patient or laboratory virus-derived PCR fragments and the corresponding NL4-3 vector where the whole Gag-PR, RT-RNaseH or IN coding regions has been deleted through inverse PCR. The susceptibility to nucleoside (NRTIs) and non-nucleoside (NNRTIs) RT inhibitors and to IN inhibitors (INIs) is calculated through a single-round infection assay in TZM-bl cells, while protease inhibitor (PI) activity is determined through a first round of infection in MT-2 cells followed by infection of TZM-bl cells with MT-2 supernatants.
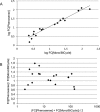
Results: The assay showed excellent reproducibility and accuracy when testing PI, NRTI, NNRTI, and INI susceptibility of drug-resistant clones previously characterized through the reference pseudoparticle-based Phenosense assay. The coefficient of interassay variation in fold change (FC) resistance was 12.0%-24.3% when assaying seven drug/clones pairs in three runs. FC values calculated by the Phenosense and in-house for 20 drug/clones pairs were in good agreement, with mean±SD ratio of 1.14±0.33 and no cases differing by more than twofold.
Conclusions: The described phenotypic assay can be adopted to evaluate the antiviral activity of licensed and investigational HIV-1 drugs targeting any of the three HIV-1 enzymes.
Keywords: HIV-1; HIV-1 inhibitors; drug resistance; phenotypic assay.
© 2017 Wiley Periodicals, Inc.
Figures

Similar articles
-
Novel method for simultaneous quantification of phenotypic resistance to maturation, protease, reverse transcriptase, and integrase HIV inhibitors based on 3'Gag(p2/p7/p1/p6)/PR/RT/INT-recombinant viruses: a useful tool in the multitarget era of antiretroviral therapy.Antimicrob Agents Chemother. 2011 Aug;55(8):3729-42. doi: 10.1128/AAC.00396-11. Epub 2011 May 31. Antimicrob Agents Chemother. 2011. PMID: 21628544 Free PMC article.
-
Comparison of susceptibility of HIV-1 variants to antiretroviral drugs by genotypic and recombinant virus phenotypic analyses.Int J Infect Dis. 2015 Aug;37:86-92. doi: 10.1016/j.ijid.2015.06.011. Epub 2015 Jun 24. Int J Infect Dis. 2015. PMID: 26117343
-
Phenotypic analysis of the sensitivity of HIV-1 to inhibitors of the reverse transcriptase, protease, and integrase using a self-inactivating virus vector system.J Med Virol. 2001 Jul;64(3):223-31. doi: 10.1002/jmv.1040. J Med Virol. 2001. PMID: 11424108
-
Study of the impact of HIV genotypic drug resistance testing on therapy efficacy.Verh K Acad Geneeskd Belg. 2001;63(5):447-73. Verh K Acad Geneeskd Belg. 2001. PMID: 11813503 Review.
-
Resistance of human immunodeficiency virus type 1 to reverse transcriptase and protease inhibitors: genotypic and phenotypic testing.J Clin Virol. 2001 Jun;21(3):197-212. doi: 10.1016/s1386-6532(00)00163-3. J Clin Virol. 2001. PMID: 11397656 Review.
Cited by
-
Identification of novel 2-benzoxazolinone derivatives with specific inhibitory activity against the HIV-1 nucleocapsid protein.Eur J Med Chem. 2018 Feb 10;145:154-164. doi: 10.1016/j.ejmech.2017.12.073. Epub 2017 Dec 24. Eur J Med Chem. 2018. PMID: 29324338 Free PMC article.
-
5,6-Dihydroxypyrimidine Scaffold to Target HIV-1 Nucleocapsid Protein.ACS Med Chem Lett. 2020 Mar 19;11(5):766-772. doi: 10.1021/acsmedchemlett.9b00608. eCollection 2020 May 14. ACS Med Chem Lett. 2020. PMID: 32435383 Free PMC article.
-
Synthesis and Evaluation of Bifunctional Aminothiazoles as Antiretrovirals Targeting the HIV-1 Nucleocapsid Protein.ACS Med Chem Lett. 2018 Dec 7;10(4):463-468. doi: 10.1021/acsmedchemlett.8b00506. eCollection 2019 Apr 11. ACS Med Chem Lett. 2018. PMID: 30996780 Free PMC article.
-
Anti-HIV-1 Activity of Lactic Acid in Human Cervicovaginal Fluid.mSphere. 2018 Jul 5;3(4):e00055-18. doi: 10.1128/mSphere.00055-18. mSphere. 2018. PMID: 29976641 Free PMC article.
-
In vitro cross-resistance to doravirine in a panel of HIV-1 clones harbouring multiple NNRTI resistance mutations.J Antimicrob Chemother. 2021 Jan 1;76(1):130-134. doi: 10.1093/jac/dkaa401. J Antimicrob Chemother. 2021. PMID: 32974670 Free PMC article.
References
-
- Egger M, Johnson LF. Estimating trends in life expectancy in HIV‐positive individuals. Lancet Glob Health. 2015;3:e122‐e123. - PubMed
-
- Geretti AM, Tsakiroglou M. HIV: new drugs, new guidelines. Curr Opin Infect Dis. 2014;27:545‐553. - PubMed
-
- Vandamme AM, Camacho RJ, Ceccherini‐Silberstein F, et al. European recommendations for the clinical use of HIV drug resistance testing: 2011 update. AIDS Rev. 2011;13:77‐108. - PubMed
-
- Zazzi M, Cozzi‐Lepri A, Prosperi MC. Computer‐aided optimization of combined anti‐retroviral therapy for HIV: new drugs, new drug targets and drug resistance. Curr HIV Res. 2016;14:101‐109. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

