Building a More Predictive Protein Force Field: A Systematic and Reproducible Route to AMBER-FB15
- PMID: 28306259
- PMCID: PMC9724927
- DOI: 10.1021/acs.jpcb.7b02320
Building a More Predictive Protein Force Field: A Systematic and Reproducible Route to AMBER-FB15
Abstract
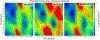
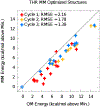
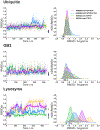
The increasing availability of high-quality experimental data and first-principles calculations creates opportunities for developing more accurate empirical force fields for simulation of proteins. We developed the AMBER-FB15 protein force field by building a high-quality quantum chemical data set consisting of comprehensive potential energy scans and employing the ForceBalance software package for parameter optimization. The optimized potential surface allows for more significant thermodynamic fluctuations away from local minima. In validation studies where simulation results are compared to experimental measurements, AMBER-FB15 in combination with the updated TIP3P-FB water model predicts equilibrium properties with equivalent accuracy, and temperature dependent properties with significantly improved accuracy, in comparison with published models. We also discuss the effect of changing the protein force field and water model on the simulation results.
Figures



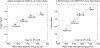
References
-
- Bash PA; Field MJ; Davenport RC; Petsko GA; Ringe D; Karplus M, Computer Simulation and Analysis of the Reaction Pathway of Triosephosphate Isomerase. Biochemistry-Us 1991, 30 (24), 5826–5832. - PubMed
-
- Gao J, Catalysis by Enzyme Conformational Change as Illustrated by Orotidine 5′-Monophosphate Decarboxylase. Current Opinion in Structural Biology 2003, 13 (2), 184–192. - PubMed
-
- Ridder L; Mulholland AJ; Rietjens IMCM; Vervoort J, A Quantum Mechanical/Molecular Mechanical Study of the Hydroxylation of Phenol and Halogenated Derivatives by Phenol Hydroxylase. J. Am. Chem. Soc. 2000, 122 (36), 8728–8738.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

