Comparative evaluation of the Minimally-Invasive Karyotyping (MINK) algorithm for non-invasive prenatal testing
- PMID: 28306738
- PMCID: PMC5356998
- DOI: 10.1371/journal.pone.0171882
Comparative evaluation of the Minimally-Invasive Karyotyping (MINK) algorithm for non-invasive prenatal testing
Abstract
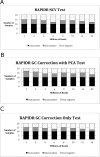
Minimally Invasive Karyotyping (MINK) was communicated in 2009 as a novel method for the non-invasive detection of fetal copy number anomalies in maternal plasma DNA. The original manuscript illustrated the potential of MINK using a model system in which fragmented genomic DNA obtained from a trisomy 21 male individual was mixed with that of his karyotypically normal mother at dilutions representing fetal fractions found in maternal plasma. Although it has been previously shown that MINK is able to non-invasively detect fetal microdeletions, its utility for aneuploidy detection in maternal plasma has not previously been demonstrated. The current study illustrates the ability of MINK to detect common aneuploidy in early gestation, compares its performance to other published third party methods (and related software packages) for prenatal aneuploidy detection and evaluates the performance of these methods across a range of sequencing read inputs. Plasma samples were obtained from 416 pregnant women between gestational weeks 8.1 and 34.4. Shotgun DNA sequencing was performed and data analyzed using MINK RAPIDR and WISECONDOR. MINK performed with greater accuracy than RAPIDR and WISECONDOR, correctly identifying 60 out of 61 true trisomy cases, and reporting only one false positive in 355 normal pregnancies. Significantly, MINK achieved accurate detection of trisomy 21 using just 2 million aligned input reads, whereas WISECONDOR required 6 million reads and RAPIDR did not achieve complete accuracy at any read input tested. In conclusion, we demonstrate that MINK provides an analysis pipeline for the detection of fetal aneuploidy in samples of maternal plasma DNA.
Conflict of interest statement
Figures


References
-
- Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(42):16266–71. Epub 2008/10/08. PubMed Central PMCID: PMC2562413. 10.1073/pnas.0808319105 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

