Common nonmutational NOTCH1 activation in chronic lymphocytic leukemia
- PMID: 28314854
- PMCID: PMC5389283
- DOI: 10.1073/pnas.1702564114
Common nonmutational NOTCH1 activation in chronic lymphocytic leukemia
Abstract
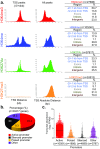
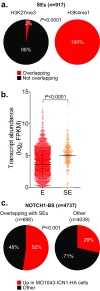
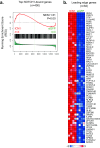
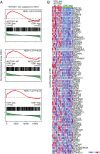
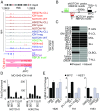
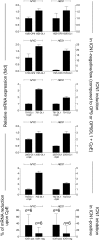
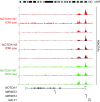
Activating mutations of NOTCH1 (a well-known oncogene in T-cell acute lymphoblastic leukemia) are present in ∼4-13% of chronic lymphocytic leukemia (CLL) cases, where they are associated with disease progression and chemorefractoriness. However, the specific role of NOTCH1 in leukemogenesis remains to be established. Here, we report that the active intracellular portion of NOTCH1 (ICN1) is detectable in ∼50% of peripheral blood CLL cases lacking gene mutations. We identify a "NOTCH1 gene-expression signature" in CLL cells, and show that this signature is significantly enriched in primary CLL cases expressing ICN1, independent of NOTCH1 mutation. NOTCH1 target genes include key regulators of B-cell proliferation, survival, and signal transduction. In particular, we show that NOTCH1 transactivates MYC via binding to B-cell-specific regulatory elements, thus implicating this oncogene in CLL development. These results significantly extend the role of NOTCH1 in CLL pathogenesis, and have direct implications for specific therapeutic targeting.
Keywords: NOTCH1; chronic lymphocytic leukemia; transcriptional network.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
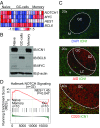
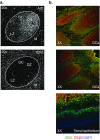
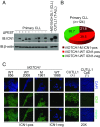

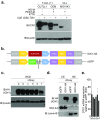



References
-
- Fabbri G, Dalla-Favera R. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat Rev Cancer. 2016;16(3):145–162. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

