Fail-safe control of translation initiation by dissociation of eIF2α phosphorylated ternary complexes
- PMID: 28315520
- PMCID: PMC5404910
- DOI: 10.7554/eLife.24542
Fail-safe control of translation initiation by dissociation of eIF2α phosphorylated ternary complexes
Abstract
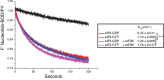
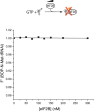
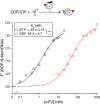
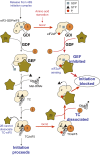
Phosphorylation of eIF2α controls translation initiation by restricting the levels of active eIF2-GTP/Met-tRNAi ternary complexes (TC). This modulates the expression of all eukaryotic mRNAs and contributes to the cellular integrated stress response. Key to controlling the activity of eIF2 are translation factors eIF2B and eIF5, thought to primarily function with eIF2-GDP and TC respectively. Using a steady-state kinetics approach with purified proteins we demonstrate that eIF2B binds to eIF2 with equal affinity irrespective of the presence or absence of competing guanine nucleotides. We show that eIF2B can compete with Met-tRNAi for eIF2-GTP and can destabilize TC. When TC is formed with unphosphorylated eIF2, eIF5 can out-compete eIF2B to stabilize TC/eIF5 complexes. However when TC/eIF5 is formed with phosphorylated eIF2, eIF2B outcompetes eIF5 and destabilizes TC. These data uncover competition between eIF2B and eIF5 for TC and identify that phosphorylated eIF2-GTP translation initiation intermediate complexes can be inhibited by eIF2B.
Keywords: ISR; S. cerevisiae; biochemistry; chromosomes; eIF2; eIF2B; genes; protein interactions; translational control.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures






Comment in
-
An emergency brake for protein synthesis.Elife. 2017 Apr 25;6:e27085. doi: 10.7554/eLife.27085. Elife. 2017. PMID: 28440747 Free PMC article.
References
-
- Amberg DC, Burke DJ, Strathern JN. Methods in Yeast Genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2005.
-
- Asano K, Krishnamoorthy T, Phan L, Pavitt GD, Hinnebusch AG. Conserved bipartite motifs in yeast eIF5 and eIF2Bepsilon, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. The EMBO Journal. 1999;18:1673–1688. doi: 10.1093/emboj/18.6.1673. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

