Expression of 16 Nitrogenase Proteins within the Plant Mitochondrial Matrix
- PMID: 28316608
- PMCID: PMC5334340
- DOI: 10.3389/fpls.2017.00287
Expression of 16 Nitrogenase Proteins within the Plant Mitochondrial Matrix
Abstract
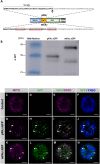
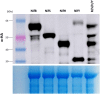
The industrial production and use of nitrogenous fertilizer involves significant environmental and economic costs. Strategies to reduce fertilizer dependency are required to address the world's increasing demand for sustainable food, fibers, and biofuels. Biological nitrogen fixation, a process unique to diazatrophic bacteria, is catalyzed by the nitrogenase complex, and reconstituting this function in plant cells is an ambitious biotechnological strategy to reduce fertilizer use. Here we establish that the full array of biosynthetic and catalytic nitrogenase (Nif) proteins from the diazotroph Klebsiella pneumoniae can be individually expressed as mitochondrial targeting peptide (MTP)-Nif fusions in Nicotiana benthamiana. We show that these are correctly targeted to the plant mitochondrial matrix, a subcellular location with biochemical and genetic characteristics potentially supportive of nitrogenase function. Although Nif proteins B, D, E, F, H, J, K, M, N, Q, S, U, V, X, Y, and Z were all detectable by Western blot analysis, the NifD catalytic component was the least abundant. To address this problem, a translational fusion between NifD and NifK was designed based on the crystal structure of the nitrogenase MoFe protein heterodimer. This fusion protein enabled equimolar NifD:NifK stoichiometry and improved NifD expression levels in plants. Finally, four MTP-Nif fusion proteins (B, S, H, Y) were successfully co-expressed, demonstrating that multiple components of nitrogenase can be targeted to plant mitochondria. These results establish the feasibility of reconstituting the complete componentry for nitrogenase in plant cells, within an intracellular environment that could support the conversion of nitrogen gas into ammonia.
Keywords: metabolic engineering; mitochondrial targeting; nitrogen fixation; nitrogenase; synthetic biology.
Figures




References
-
- Case D. A., Darden T. A., Cheatham T. E., Simmerling C. L., Wang J., Duke R. E., et al. (2012). AMBER 12. San Francisco, CA: University of California.
LinkOut - more resources
Full Text Sources
Other Literature Sources

