DNA damage during S-phase mediates the proliferation-quiescence decision in the subsequent G1 via p21 expression
- PMID: 28317845
- PMCID: PMC5364389
- DOI: 10.1038/ncomms14728
DNA damage during S-phase mediates the proliferation-quiescence decision in the subsequent G1 via p21 expression
Abstract
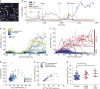
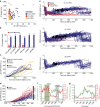
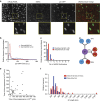
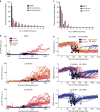
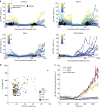
Following DNA damage caused by exogenous sources, such as ionizing radiation, the tumour suppressor p53 mediates cell cycle arrest via expression of the CDK inhibitor, p21. However, the role of p21 in maintaining genomic stability in the absence of exogenous DNA-damaging agents is unclear. Here, using live single-cell measurements of p21 protein in proliferating cultures, we show that naturally occurring DNA damage incurred over S-phase causes p53-dependent accumulation of p21 during mother G2- and daughter G1-phases. High p21 levels mediate G1 arrest via CDK inhibition, yet lower levels have no impact on G1 progression, and the ubiquitin ligases CRL4Cdt2 and SCFSkp2 couple to degrade p21 prior to the G1/S transition. Mathematical modelling reveals that a bistable switch, created by CRL4Cdt2, promotes irreversible S-phase entry by keeping p21 levels low, preventing premature S-phase exit upon DNA damage. Thus, we characterize how p21 regulates the proliferation-quiescence decision to maintain genomic stability.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Jeggo P. A., Pearl L. H. & Carr A. M. DNA repair, genome stability and cancer: a historical perspective. Nat. Rev. Cancer 16, 35–42 (2016). - PubMed
-
- Kastan M. B., Onyekwere O., Sidransky D., Vogelstein B. & Craig R. W. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 51, 6304–6311 (1991). - PubMed
-
- Lane D. P. & Crawford L. V. T antigen is bound to a host protein in SY40-transformed cells. Nature 278, 261–263 (1979). - PubMed
-
- el-Deiry W. S. et al. WAF1, a potential mediator of p53 tumor suppression. Cell 75, 817–825 (1993). - PubMed
-
- el-Deiry W. S. et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 54, 1169–1174 (1994). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

