Structural and functional studies of a noncanonical Dicer from Entamoeba histolytica
- PMID: 28317870
- PMCID: PMC5357909
- DOI: 10.1038/srep44832
Structural and functional studies of a noncanonical Dicer from Entamoeba histolytica
Abstract
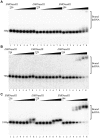
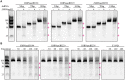
RNaseIII proteins are dsRNA-specific endonucleases involved in many important biological processes, such as small RNA processing and maturation in eukaryotes. Various small RNAs have been identified in a protozoan parasite Entamoeba histolytica. EhRNaseIII is the only RNaseIII endonuclease domain (RIIID)-containing protein in E. histolytica. Here, we present three crystal structures that reveal several unique structural features of EhRNaseIII, especially the interactions between the two helixes (α1 and α7) flanking the RIIID core domain. Structure and sequence analysis indicate that EhRNaseIII is a noncanonical Dicer and it lacks a dsRBD in the C-terminal region (CTR). In vitro studies suggest that EhRNaseIII prefers to bind and cleave longer dsRNAs, generating products around 25 nucleotides in length. Truncation of the CTR or attaching the dsRBD of Aquifex aeolicus RNaseIII can enhance the binding and cleavage activities of EhRNaseIII. In combination with in vitro crosslinking assay, our results suggested that EhRNaseIII functions in a cooperative mode. We speculate that some partner proteins may exist in E. histolytica and regulates the activity of EhRNaseIII through interaction with its CTR. Our studies support that EhRNaseIII plays an important role in producing small RNAs in E. histolytica.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





References
-
- Hutvagner G. & Zamore P. D. RNAi: nature abhors a double-strand. Curr Opin Genet Dev 12, 225–232 (2002). - PubMed
-
- Nicholson A. W. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol Rev 23, 371–390 (1999). - PubMed
-
- MacRae I. J., Zhou K. & Doudna J. A. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol 14, 934–940 (2007). - PubMed
-
- Kawamata T. & Tomari Y. Making RISC. Trends Biochem Sci 35, 368–376 (2010). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

